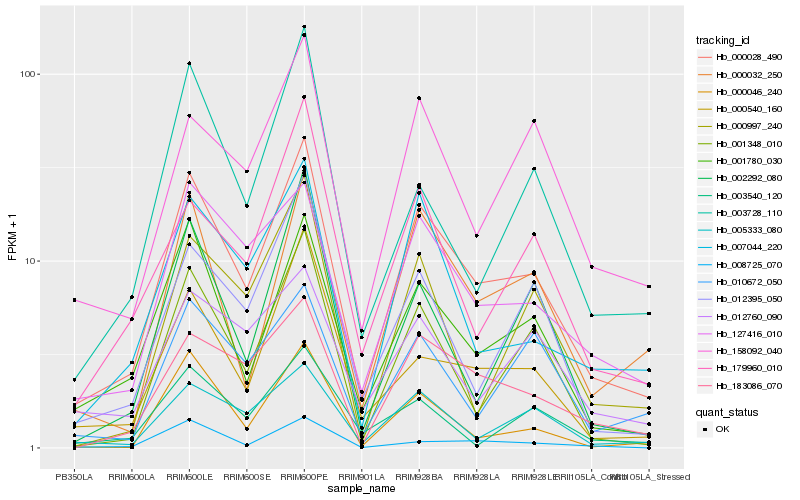
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000028_490 |
0.0 |
- |
- |
calmodulin-like protein 6a [Populus trichocarpa] |
| 2 |
Hb_001780_030 |
0.1198191646 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At5g41260 [Jatropha curcas] |
| 3 |
Hb_127416_010 |
0.1389382963 |
- |
- |
LysM domain GPI-anchored protein 2 precursor, putative [Ricinus communis] |
| 4 |
Hb_183086_070 |
0.1432074654 |
- |
- |
PREDICTED: mitochondrial chaperone BCS1-like [Jatropha curcas] |
| 5 |
Hb_005333_080 |
0.1438124119 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000032_250 |
0.1445732334 |
- |
- |
PREDICTED: endoglucanase 6 [Jatropha curcas] |
| 7 |
Hb_007044_220 |
0.1457802991 |
- |
- |
Allene oxide cyclase 4, chloroplast precursor, putative [Ricinus communis] |
| 8 |
Hb_001348_010 |
0.1467649923 |
- |
- |
PREDICTED: lysM domain-containing GPI-anchored protein 1 [Jatropha curcas] |
| 9 |
Hb_000540_160 |
0.1526047849 |
- |
- |
PREDICTED: mitogen-activated protein kinase homolog NTF6 [Jatropha curcas] |
| 10 |
Hb_012395_050 |
0.157106838 |
- |
- |
PREDICTED: uncharacterized protein LOC105632180 [Jatropha curcas] |
| 11 |
Hb_000046_240 |
0.1579635602 |
- |
- |
PREDICTED: uncharacterized protein LOC105631837 [Jatropha curcas] |
| 12 |
Hb_158092_040 |
0.1598028748 |
- |
- |
PREDICTED: uncharacterized protein LOC105647294 [Jatropha curcas] |
| 13 |
Hb_179960_010 |
0.1660643109 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase ATL4-like [Jatropha curcas] |
| 14 |
Hb_000997_240 |
0.1704869803 |
transcription factor |
TF Family: AUX/IAA |
Auxin-responsive protein IAA4, putative [Ricinus communis] |
| 15 |
Hb_012760_090 |
0.1716244557 |
- |
- |
Aspartic proteinase-like protein 1 [Morus notabilis] |
| 16 |
Hb_010672_050 |
0.1728646813 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 2 isoform X3 [Jatropha curcas] |
| 17 |
Hb_002292_080 |
0.1729038619 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 18 |
Hb_003728_110 |
0.1744872016 |
- |
- |
PREDICTED: calmodulin-like protein 3 [Populus euphratica] |
| 19 |
Hb_008725_070 |
0.1747084531 |
- |
- |
Cellulose synthase A catalytic subunit 3 [UDP-forming], putative [Ricinus communis] |
| 20 |
Hb_003540_120 |
0.1757099614 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |