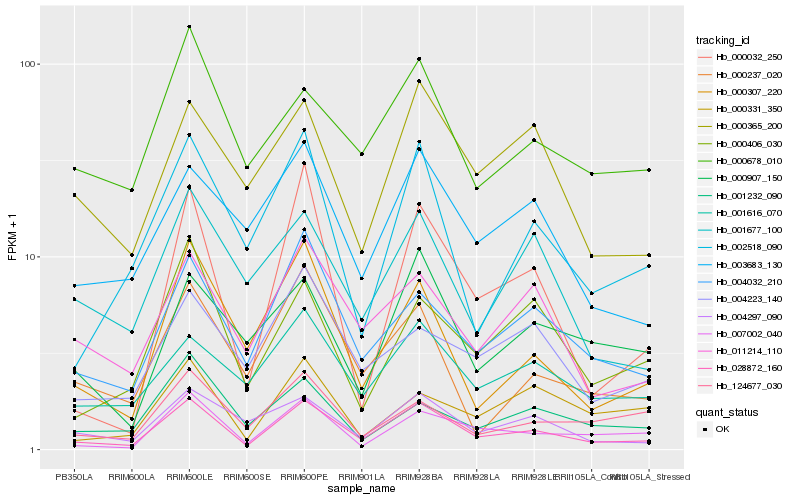
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_028872_160 |
0.0 |
- |
- |
PREDICTED: ATP-dependent DNA helicase Q-like 5 [Jatropha curcas] |
| 2 |
Hb_004032_210 |
0.1487820061 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_007002_040 |
0.1544644871 |
transcription factor |
TF Family: PHD |
DNA replication regulator dpb11, putative [Ricinus communis] |
| 4 |
Hb_000307_220 |
0.1614430571 |
- |
- |
oxidoreductase family protein [Populus trichocarpa] |
| 5 |
Hb_000032_250 |
0.1640230084 |
- |
- |
PREDICTED: endoglucanase 6 [Jatropha curcas] |
| 6 |
Hb_011214_110 |
0.1664864057 |
- |
- |
PREDICTED: ALA-interacting subunit 3-like [Jatropha curcas] |
| 7 |
Hb_002518_090 |
0.1701952318 |
- |
- |
PREDICTED: tubulin beta-2 chain [Jatropha curcas] |
| 8 |
Hb_004297_090 |
0.1703989125 |
- |
- |
PREDICTED: DNA polymerase epsilon catalytic subunit A [Jatropha curcas] |
| 9 |
Hb_001616_070 |
0.1709522346 |
- |
- |
PREDICTED: uncharacterized protein LOC105644365 [Jatropha curcas] |
| 10 |
Hb_000406_030 |
0.1746834099 |
- |
- |
Bifunctional 3-dehydroquinate dehydratase/shikimate dehydrogenase [Morus notabilis] |
| 11 |
Hb_003683_130 |
0.1785780452 |
- |
- |
PREDICTED: enolase 1, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000237_020 |
0.1787792181 |
- |
- |
PREDICTED: DNA replication licensing factor MCM4 [Jatropha curcas] |
| 13 |
Hb_000365_200 |
0.1796097169 |
- |
- |
PREDICTED: 2,3-bisphosphoglycerate-independent phosphoglycerate mutase [Jatropha curcas] |
| 14 |
Hb_000678_010 |
0.1808873955 |
- |
- |
PREDICTED: ketol-acid reductoisomerase, chloroplastic-like [Jatropha curcas] |
| 15 |
Hb_000907_150 |
0.1850030854 |
- |
- |
hypothetical protein L484_004295 [Morus notabilis] |
| 16 |
Hb_001677_100 |
0.1860943311 |
- |
- |
delta1-pyrroline-5-carboxylate synthase [Manihot esculenta] |
| 17 |
Hb_004223_140 |
0.1869018003 |
- |
- |
endo-1,4-beta-glucanase, putative [Ricinus communis] |
| 18 |
Hb_124677_030 |
0.1882848544 |
- |
- |
PREDICTED: uncharacterized protein LOC105650473 [Jatropha curcas] |
| 19 |
Hb_001232_090 |
0.189857718 |
- |
- |
PREDICTED: regulator of telomere elongation helicase 1 homolog [Jatropha curcas] |
| 20 |
Hb_000331_350 |
0.1906871906 |
- |
- |
hypothetical protein JCGZ_11337 [Jatropha curcas] |