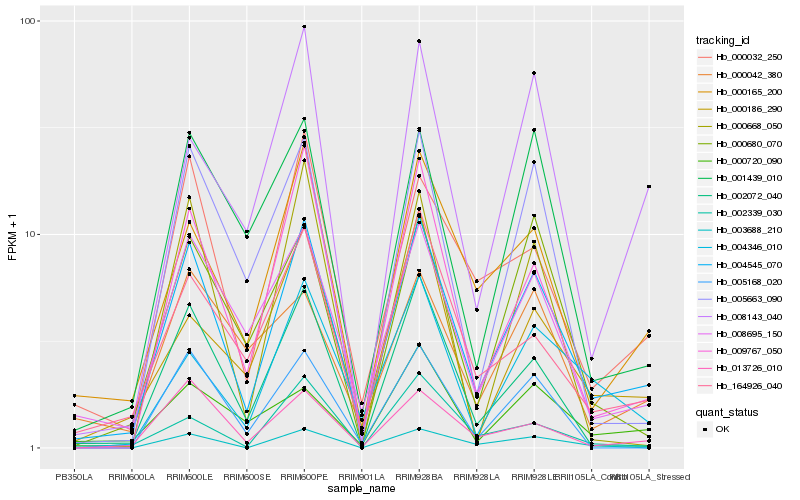
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004545_070 |
0.0 |
- |
- |
PREDICTED: bifunctional 3-dehydroquinate dehydratase/shikimate dehydrogenase, chloroplastic-like [Jatropha curcas] |
| 2 |
Hb_008695_150 |
0.1352079841 |
- |
- |
PREDICTED: beta-1,3-galactosyltransferase 7 isoform X1 [Jatropha curcas] |
| 3 |
Hb_009767_050 |
0.1420719628 |
- |
- |
PREDICTED: uncharacterized protein LOC105631978 isoform X1 [Jatropha curcas] |
| 4 |
Hb_013726_010 |
0.1436086352 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 5 |
Hb_000165_200 |
0.1447159065 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 6 |
Hb_000032_250 |
0.1489687361 |
- |
- |
PREDICTED: endoglucanase 6 [Jatropha curcas] |
| 7 |
Hb_002072_040 |
0.1515492388 |
- |
- |
PREDICTED: uncharacterized protein LOC105630881 [Jatropha curcas] |
| 8 |
Hb_164926_040 |
0.1586194345 |
- |
- |
Beta-1,3-galactosyltransferase sqv-2, putative [Ricinus communis] |
| 9 |
Hb_001439_010 |
0.1667610198 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucosyltransferase 5-like [Jatropha curcas] |
| 10 |
Hb_005663_090 |
0.1670375299 |
- |
- |
PREDICTED: alcohol dehydrogenase-like 7 [Jatropha curcas] |
| 11 |
Hb_005168_020 |
0.1708996295 |
- |
- |
PREDICTED: U-box domain-containing protein 30-like [Jatropha curcas] |
| 12 |
Hb_000680_070 |
0.1744986768 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase ALE2 isoform X1 [Jatropha curcas] |
| 13 |
Hb_003688_210 |
0.1746599512 |
- |
- |
replication factor A 1, rfa1, putative [Ricinus communis] |
| 14 |
Hb_004346_010 |
0.1756113852 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000720_090 |
0.1825809806 |
transcription factor |
TF Family: MYB |
myb, putative [Ricinus communis] |
| 16 |
Hb_000186_290 |
0.1833656711 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 8 isoform X1 [Jatropha curcas] |
| 17 |
Hb_002339_030 |
0.1840472477 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 18 |
Hb_008143_040 |
0.1845344697 |
- |
- |
Alpha-expansin 4 precursor, putative [Ricinus communis] |
| 19 |
Hb_000668_050 |
0.188217269 |
- |
- |
phosphoglycerate mutase, putative [Ricinus communis] |
| 20 |
Hb_000042_380 |
0.188471768 |
- |
- |
PREDICTED: uncharacterized membrane protein At3g27390 [Jatropha curcas] |