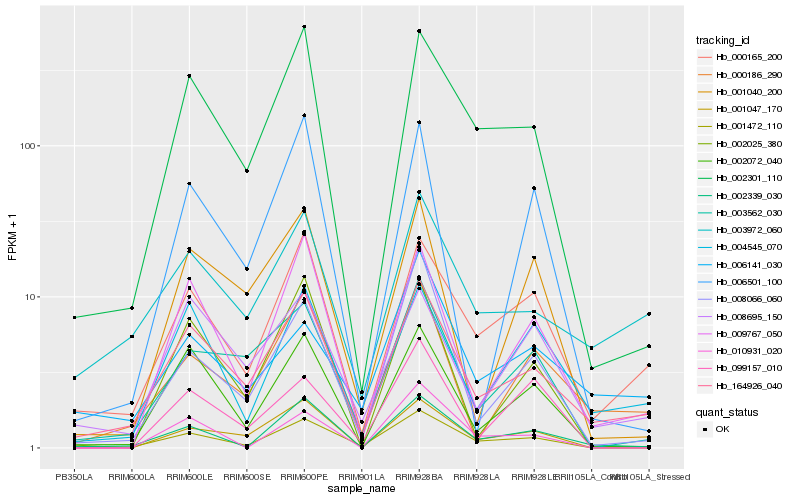
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001472_110 |
0.0 |
desease resistance |
Gene Name: TIR_2 |
nucleoside-triphosphatase, putative [Ricinus communis] |
| 2 |
Hb_164926_040 |
0.1352554381 |
- |
- |
Beta-1,3-galactosyltransferase sqv-2, putative [Ricinus communis] |
| 3 |
Hb_000165_200 |
0.1361290146 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 4 |
Hb_002301_110 |
0.1364224746 |
- |
- |
PREDICTED: protein E6-like [Populus euphratica] |
| 5 |
Hb_002339_030 |
0.1527392792 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 6 |
Hb_003562_030 |
0.1700594831 |
- |
- |
- |
| 7 |
Hb_002072_040 |
0.1739168605 |
- |
- |
PREDICTED: uncharacterized protein LOC105630881 [Jatropha curcas] |
| 8 |
Hb_008066_060 |
0.174706616 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: subtilisin-like protease SBT1.6 [Jatropha curcas] |
| 9 |
Hb_001047_170 |
0.1828971536 |
- |
- |
PREDICTED: uncharacterized protein LOC105649981 [Jatropha curcas] |
| 10 |
Hb_009767_050 |
0.1845929912 |
- |
- |
PREDICTED: uncharacterized protein LOC105631978 isoform X1 [Jatropha curcas] |
| 11 |
Hb_003972_060 |
0.1949901886 |
- |
- |
PREDICTED: UDP-arabinopyranose mutase 3 [Populus euphratica] |
| 12 |
Hb_004545_070 |
0.1959519861 |
- |
- |
PREDICTED: bifunctional 3-dehydroquinate dehydratase/shikimate dehydrogenase, chloroplastic-like [Jatropha curcas] |
| 13 |
Hb_010931_020 |
0.2004082799 |
- |
- |
Kunitz-type protease inhibitor KPI-F4 [Populus trichocarpa x Populus deltoides] |
| 14 |
Hb_000186_290 |
0.2015875295 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 8 isoform X1 [Jatropha curcas] |
| 15 |
Hb_006141_030 |
0.2077448832 |
- |
- |
- |
| 16 |
Hb_099157_010 |
0.2102816032 |
- |
- |
PREDICTED: uncharacterized protein LOC105630881 [Jatropha curcas] |
| 17 |
Hb_002025_380 |
0.2103090301 |
- |
- |
Uncharacterized protein TCM_011176 [Theobroma cacao] |
| 18 |
Hb_008695_150 |
0.211972636 |
- |
- |
PREDICTED: beta-1,3-galactosyltransferase 7 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001040_200 |
0.2131130372 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA1H1-like [Jatropha curcas] |
| 20 |
Hb_006501_100 |
0.2161287002 |
- |
- |
malic enzyme, putative [Ricinus communis] |