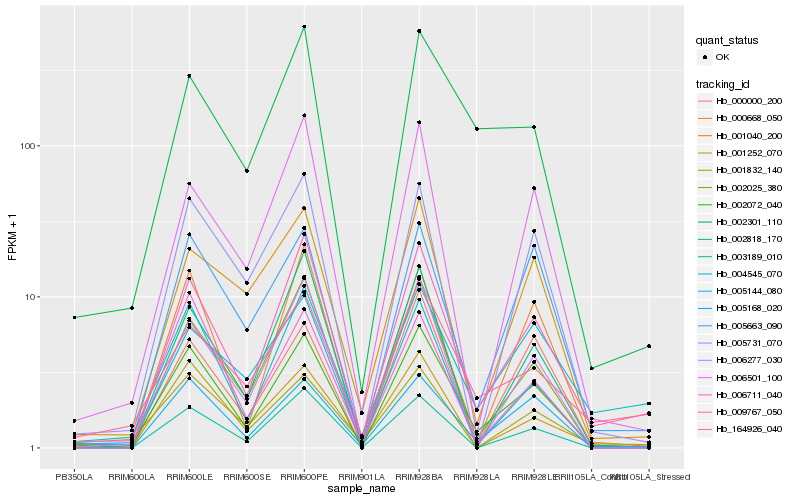
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002072_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105630881 [Jatropha curcas] |
| 2 |
Hb_002025_380 |
0.1170100804 |
- |
- |
Uncharacterized protein TCM_011176 [Theobroma cacao] |
| 3 |
Hb_009767_050 |
0.1173678327 |
- |
- |
PREDICTED: uncharacterized protein LOC105631978 isoform X1 [Jatropha curcas] |
| 4 |
Hb_005168_020 |
0.1239164639 |
- |
- |
PREDICTED: U-box domain-containing protein 30-like [Jatropha curcas] |
| 5 |
Hb_006277_030 |
0.1307178667 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_003189_010 |
0.1350789314 |
- |
- |
hypothetical protein EUGRSUZ_G02605 [Eucalyptus grandis] |
| 7 |
Hb_005731_070 |
0.140531653 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_005663_090 |
0.1425429123 |
- |
- |
PREDICTED: alcohol dehydrogenase-like 7 [Jatropha curcas] |
| 9 |
Hb_005144_080 |
0.1465406873 |
- |
- |
Serine carboxypeptidase, putative [Ricinus communis] |
| 10 |
Hb_002301_110 |
0.1470030882 |
- |
- |
PREDICTED: protein E6-like [Populus euphratica] |
| 11 |
Hb_000668_050 |
0.1481602754 |
- |
- |
phosphoglycerate mutase, putative [Ricinus communis] |
| 12 |
Hb_004545_070 |
0.1515492388 |
- |
- |
PREDICTED: bifunctional 3-dehydroquinate dehydratase/shikimate dehydrogenase, chloroplastic-like [Jatropha curcas] |
| 13 |
Hb_006501_100 |
0.1518135641 |
- |
- |
malic enzyme, putative [Ricinus communis] |
| 14 |
Hb_002818_170 |
0.1531754813 |
- |
- |
hypothetical protein POPTR_0014s10650g [Populus trichocarpa] |
| 15 |
Hb_001040_200 |
0.1535507008 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA1H1-like [Jatropha curcas] |
| 16 |
Hb_006711_040 |
0.1538215124 |
- |
- |
PREDICTED: probable protein ABIL5 [Jatropha curcas] |
| 17 |
Hb_001252_070 |
0.1573745631 |
- |
- |
hypothetical protein JCGZ_01385 [Jatropha curcas] |
| 18 |
Hb_001832_140 |
0.1596557166 |
- |
- |
endomembrane protein 70 family protein [Medicago truncatula] |
| 19 |
Hb_000000_200 |
0.1613676416 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12 [Jatropha curcas] |
| 20 |
Hb_164926_040 |
0.1613698572 |
- |
- |
Beta-1,3-galactosyltransferase sqv-2, putative [Ricinus communis] |