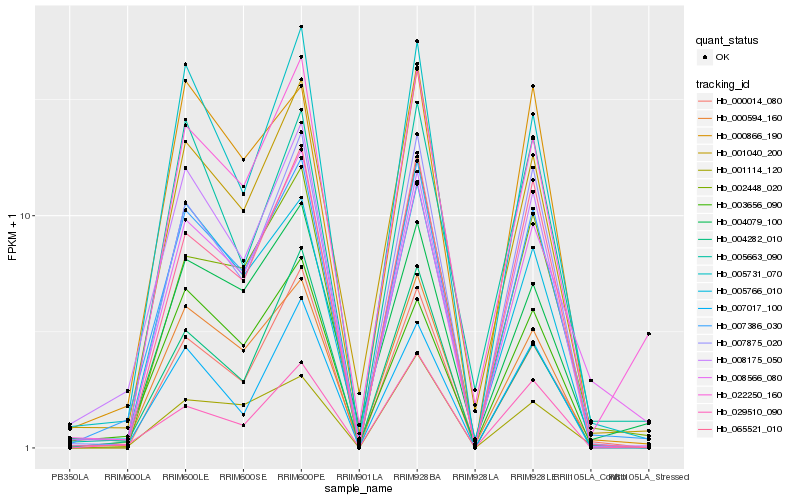
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007386_030 |
0.0 |
- |
- |
PREDICTED: formin-like protein 2 [Jatropha curcas] |
| 2 |
Hb_000594_160 |
0.079893463 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 8 [Jatropha curcas] |
| 3 |
Hb_007875_020 |
0.0830186231 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme-like [Jatropha curcas] |
| 4 |
Hb_005731_070 |
0.0884660239 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_065521_010 |
0.0918173371 |
- |
- |
PREDICTED: probable metal-nicotianamine transporter YSL5 [Jatropha curcas] |
| 6 |
Hb_001040_200 |
0.0945105496 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA1H1-like [Jatropha curcas] |
| 7 |
Hb_008566_080 |
0.0985912834 |
- |
- |
PREDICTED: uncharacterized protein LOC105647834 [Jatropha curcas] |
| 8 |
Hb_002448_020 |
0.0999140558 |
transcription factor |
TF Family: SBP |
Squamosa promoter-binding protein, putative [Ricinus communis] |
| 9 |
Hb_022250_160 |
0.0999821879 |
- |
- |
respiratory burst oxidase A [Manihot esculenta] |
| 10 |
Hb_000014_080 |
0.1013422254 |
- |
- |
PREDICTED: UDP-glycosyltransferase 87A1-like [Jatropha curcas] |
| 11 |
Hb_005766_010 |
0.1015994503 |
- |
- |
serine carboxypeptidase, putative [Ricinus communis] |
| 12 |
Hb_008175_050 |
0.1036889922 |
- |
- |
PREDICTED: formin-like protein 2 [Jatropha curcas] |
| 13 |
Hb_007017_100 |
0.1049907908 |
- |
- |
PREDICTED: uncharacterized protein LOC105649804 isoform X1 [Jatropha curcas] |
| 14 |
Hb_029510_090 |
0.1056555008 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At5g63020 [Jatropha curcas] |
| 15 |
Hb_000866_190 |
0.1082925086 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 16 |
Hb_001114_120 |
0.1112800221 |
- |
- |
protein with unknown function [Ricinus communis] |
| 17 |
Hb_004079_100 |
0.1117833796 |
- |
- |
PREDICTED: uncharacterized protein LOC105637705 [Jatropha curcas] |
| 18 |
Hb_003656_090 |
0.1174422992 |
- |
- |
PREDICTED: protein Brevis radix-like 4 [Jatropha curcas] |
| 19 |
Hb_004282_010 |
0.1179431918 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At2g24230 [Jatropha curcas] |
| 20 |
Hb_005663_090 |
0.1179681011 |
- |
- |
PREDICTED: alcohol dehydrogenase-like 7 [Jatropha curcas] |