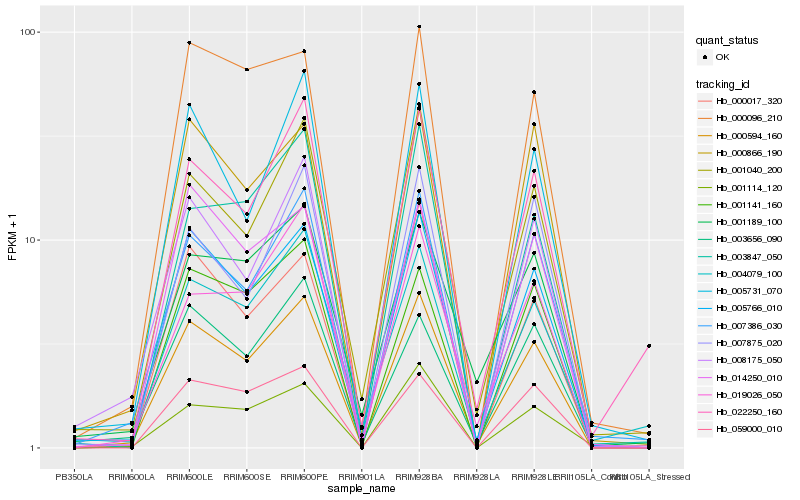
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000594_160 |
0.0 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 8 [Jatropha curcas] |
| 2 |
Hb_005766_010 |
0.0499738377 |
- |
- |
serine carboxypeptidase, putative [Ricinus communis] |
| 3 |
Hb_007386_030 |
0.079893463 |
- |
- |
PREDICTED: formin-like protein 2 [Jatropha curcas] |
| 4 |
Hb_001040_200 |
0.0885180403 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA1H1-like [Jatropha curcas] |
| 5 |
Hb_005731_070 |
0.0967559671 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_004079_100 |
0.103181381 |
- |
- |
PREDICTED: uncharacterized protein LOC105637705 [Jatropha curcas] |
| 7 |
Hb_000866_190 |
0.1057500834 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 8 |
Hb_001141_160 |
0.1077970754 |
- |
- |
PREDICTED: sugar transport protein 14-like [Jatropha curcas] |
| 9 |
Hb_022250_160 |
0.1084533283 |
- |
- |
respiratory burst oxidase A [Manihot esculenta] |
| 10 |
Hb_003656_090 |
0.1087195165 |
- |
- |
PREDICTED: protein Brevis radix-like 4 [Jatropha curcas] |
| 11 |
Hb_059000_010 |
0.1100009525 |
- |
- |
hypothetical protein JCGZ_17736 [Jatropha curcas] |
| 12 |
Hb_000096_210 |
0.1126880207 |
- |
- |
PREDICTED: probable carboxylesterase 7 [Jatropha curcas] |
| 13 |
Hb_003847_050 |
0.1127881987 |
- |
- |
PREDICTED: aquaporin SIP1-1 [Jatropha curcas] |
| 14 |
Hb_000017_320 |
0.1130735631 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor GLABRA 3-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_001114_120 |
0.1136652232 |
- |
- |
protein with unknown function [Ricinus communis] |
| 16 |
Hb_019026_050 |
0.1143490058 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_008175_050 |
0.1149608714 |
- |
- |
PREDICTED: formin-like protein 2 [Jatropha curcas] |
| 18 |
Hb_014250_010 |
0.1167540387 |
- |
- |
PREDICTED: alpha-mannosidase-like isoform X2 [Populus euphratica] |
| 19 |
Hb_007875_020 |
0.1216338321 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme-like [Jatropha curcas] |
| 20 |
Hb_001189_100 |
0.1217234472 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 2-like [Jatropha curcas] |