| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
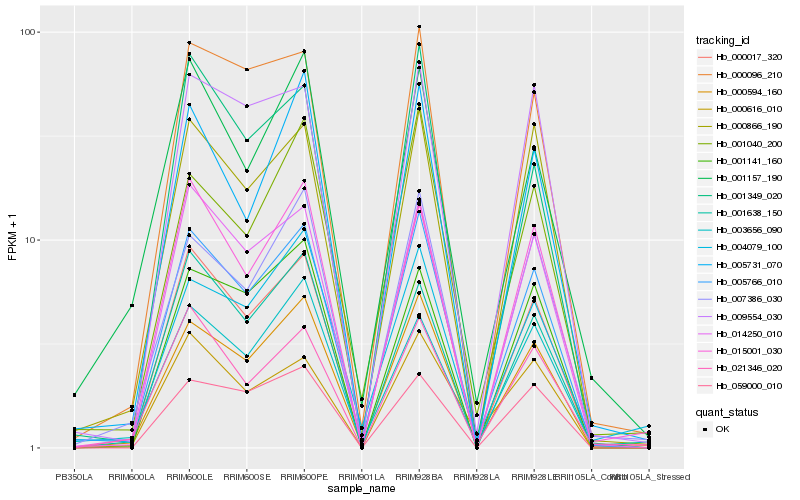
Hb_005766_010 |
0.0 |
- |
- |
serine carboxypeptidase, putative [Ricinus communis] |
| 2 |
Hb_000594_160 |
0.0499738377 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 8 [Jatropha curcas] |
| 3 |
Hb_014250_010 |
0.0859476792 |
- |
- |
PREDICTED: alpha-mannosidase-like isoform X2 [Populus euphratica] |
| 4 |
Hb_000866_190 |
0.0899967057 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 5 |
Hb_000096_210 |
0.0973967963 |
- |
- |
PREDICTED: probable carboxylesterase 7 [Jatropha curcas] |
| 6 |
Hb_000017_320 |
0.0992463536 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor GLABRA 3-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_005731_070 |
0.0996841687 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_007386_030 |
0.1015994503 |
- |
- |
PREDICTED: formin-like protein 2 [Jatropha curcas] |
| 9 |
Hb_015001_030 |
0.1044703163 |
- |
- |
inosine-uridine preferring nucleoside hydrolase family protein [Populus trichocarpa] |
| 10 |
Hb_021346_020 |
0.1051349202 |
- |
- |
hypothetical protein JCGZ_26746 [Jatropha curcas] |
| 11 |
Hb_009554_030 |
0.1089104721 |
- |
- |
PREDICTED: chlorophyll synthase, chloroplastic [Prunus mume] |
| 12 |
Hb_001040_200 |
0.1098032148 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA1H1-like [Jatropha curcas] |
| 13 |
Hb_003656_090 |
0.1105430365 |
- |
- |
PREDICTED: protein Brevis radix-like 4 [Jatropha curcas] |
| 14 |
Hb_001349_020 |
0.1109169751 |
- |
- |
PREDICTED: leucoanthocyanidin reductase-like [Jatropha curcas] |
| 15 |
Hb_001157_190 |
0.1118231018 |
- |
- |
PREDICTED: protein YLS3 [Jatropha curcas] |
| 16 |
Hb_059000_010 |
0.1145848192 |
- |
- |
hypothetical protein JCGZ_17736 [Jatropha curcas] |
| 17 |
Hb_001141_160 |
0.1166258745 |
- |
- |
PREDICTED: sugar transport protein 14-like [Jatropha curcas] |
| 18 |
Hb_000616_010 |
0.1172792337 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001638_150 |
0.1174490167 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 20 |
Hb_004079_100 |
0.1179274648 |
- |
- |
PREDICTED: uncharacterized protein LOC105637705 [Jatropha curcas] |