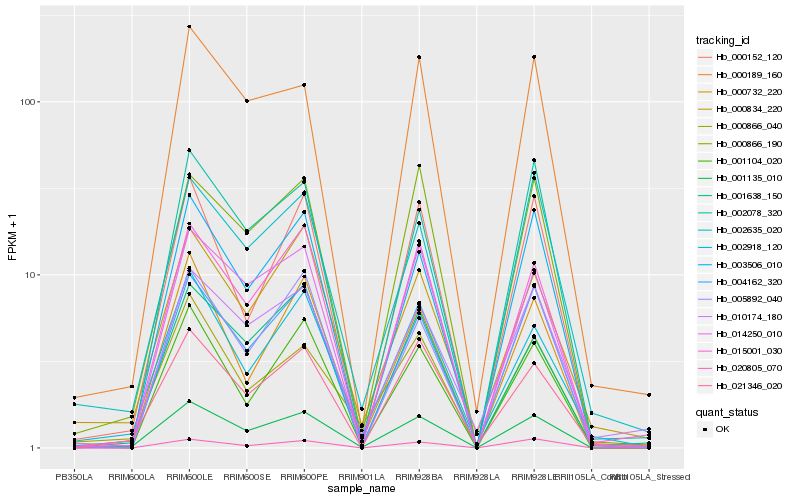
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001135_010 |
0.0 |
- |
- |
hypothetical protein CICLE_v10024612mg [Citrus clementina] |
| 2 |
Hb_004162_320 |
0.0894846349 |
- |
- |
PREDICTED: uncharacterized protein LOC105628009 isoform X1 [Jatropha curcas] |
| 3 |
Hb_015001_030 |
0.0930282038 |
- |
- |
inosine-uridine preferring nucleoside hydrolase family protein [Populus trichocarpa] |
| 4 |
Hb_003506_010 |
0.0949389047 |
- |
- |
hypothetical protein JCGZ_01146 [Jatropha curcas] |
| 5 |
Hb_014250_010 |
0.0997224477 |
- |
- |
PREDICTED: alpha-mannosidase-like isoform X2 [Populus euphratica] |
| 6 |
Hb_000866_190 |
0.1000361369 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 7 |
Hb_002918_120 |
0.1054954658 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 8 |
Hb_000189_160 |
0.1071273405 |
- |
- |
PREDICTED: GDP-mannose 3,5-epimerase 2 [Eucalyptus grandis] |
| 9 |
Hb_021346_020 |
0.1077718279 |
- |
- |
hypothetical protein JCGZ_26746 [Jatropha curcas] |
| 10 |
Hb_001104_020 |
0.1093427909 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 11 |
Hb_000732_220 |
0.111846568 |
- |
- |
PREDICTED: importin subunit alpha-5 [Jatropha curcas] |
| 12 |
Hb_000152_120 |
0.1131415768 |
- |
- |
alpha-l-fucosidase, putative [Ricinus communis] |
| 13 |
Hb_002078_320 |
0.1143427801 |
- |
- |
PREDICTED: glucuronoxylan 4-O-methyltransferase 1 [Jatropha curcas] |
| 14 |
Hb_002635_020 |
0.1144412185 |
- |
- |
hypothetical protein POPTR_0018s09790g [Populus trichocarpa] |
| 15 |
Hb_005892_040 |
0.1159667922 |
- |
- |
hypothetical protein JCGZ_02368 [Jatropha curcas] |
| 16 |
Hb_000866_040 |
0.1178196061 |
- |
- |
PREDICTED: cytochrome P450 86A8-like [Jatropha curcas] |
| 17 |
Hb_000834_220 |
0.1179759179 |
- |
- |
PREDICTED: uncharacterized protein LOC105637705 [Jatropha curcas] |
| 18 |
Hb_001638_150 |
0.1182833664 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 19 |
Hb_020805_070 |
0.1189622121 |
- |
- |
PREDICTED: MLO protein homolog 1-like [Jatropha curcas] |
| 20 |
Hb_010174_180 |
0.1193283538 |
- |
- |
protein binding protein, putative [Ricinus communis] |