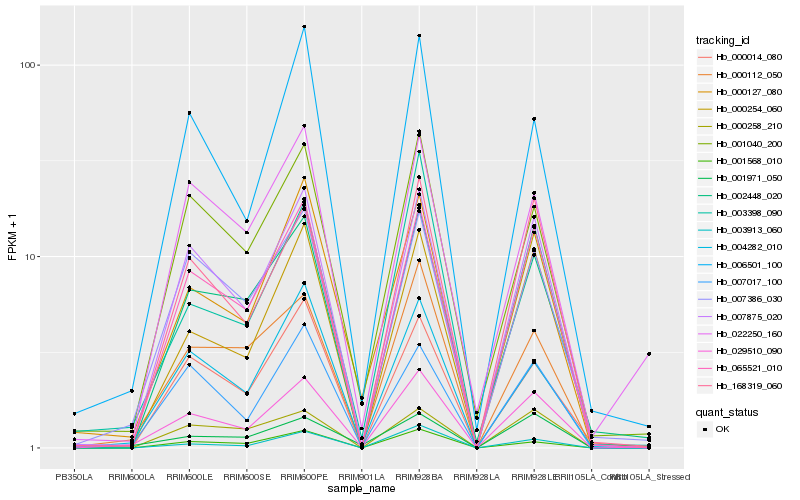
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_029510_090 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At5g63020 [Jatropha curcas] |
| 2 |
Hb_007875_020 |
0.0859273805 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme-like [Jatropha curcas] |
| 3 |
Hb_065521_010 |
0.0868257635 |
- |
- |
PREDICTED: probable metal-nicotianamine transporter YSL5 [Jatropha curcas] |
| 4 |
Hb_168319_060 |
0.0962521993 |
- |
- |
PREDICTED: cytochrome P450 CYP749A22-like [Populus euphratica] |
| 5 |
Hb_007386_030 |
0.1056555008 |
- |
- |
PREDICTED: formin-like protein 2 [Jatropha curcas] |
| 6 |
Hb_002448_020 |
0.1111118993 |
transcription factor |
TF Family: SBP |
Squamosa promoter-binding protein, putative [Ricinus communis] |
| 7 |
Hb_000254_060 |
0.120263685 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001040_200 |
0.1228656093 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA1H1-like [Jatropha curcas] |
| 9 |
Hb_000112_050 |
0.1230691345 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL1 [Jatropha curcas] |
| 10 |
Hb_007017_100 |
0.1252670241 |
- |
- |
PREDICTED: uncharacterized protein LOC105649804 isoform X1 [Jatropha curcas] |
| 11 |
Hb_003398_090 |
0.1296213768 |
- |
- |
PREDICTED: expansin-A4 [Jatropha curcas] |
| 12 |
Hb_000127_080 |
0.1298465749 |
- |
- |
hypothetical protein POPTR_0005s21760g [Populus trichocarpa] |
| 13 |
Hb_006501_100 |
0.1320808958 |
- |
- |
malic enzyme, putative [Ricinus communis] |
| 14 |
Hb_001971_050 |
0.1353991611 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 15 |
Hb_003913_060 |
0.1358768227 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 16 |
Hb_004282_010 |
0.1388457894 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At2g24230 [Jatropha curcas] |
| 17 |
Hb_000258_210 |
0.1402612823 |
- |
- |
prenyl-dependent CAAX protease, putative [Ricinus communis] |
| 18 |
Hb_001568_010 |
0.1417252449 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_022250_160 |
0.1448735005 |
- |
- |
respiratory burst oxidase A [Manihot esculenta] |
| 20 |
Hb_000014_080 |
0.1464886267 |
- |
- |
PREDICTED: UDP-glycosyltransferase 87A1-like [Jatropha curcas] |