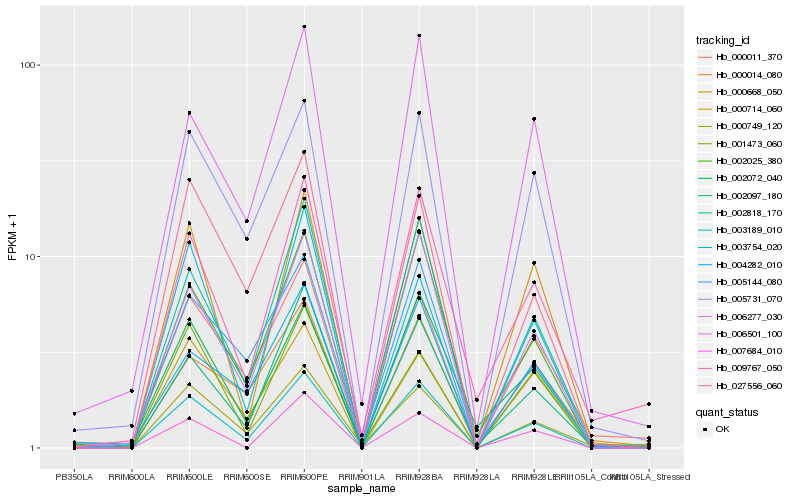
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003189_010 |
0.0 |
- |
- |
hypothetical protein EUGRSUZ_G02605 [Eucalyptus grandis] |
| 2 |
Hb_002818_170 |
0.0717552938 |
- |
- |
hypothetical protein POPTR_0014s10650g [Populus trichocarpa] |
| 3 |
Hb_006277_030 |
0.0722487764 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002025_380 |
0.0773942588 |
- |
- |
Uncharacterized protein TCM_011176 [Theobroma cacao] |
| 5 |
Hb_002097_180 |
0.1039291197 |
- |
- |
PREDICTED: probable pectate lyase 12 isoform X1 [Jatropha curcas] |
| 6 |
Hb_005731_070 |
0.1097143022 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000668_050 |
0.1164397648 |
- |
- |
phosphoglycerate mutase, putative [Ricinus communis] |
| 8 |
Hb_006501_100 |
0.1197835726 |
- |
- |
malic enzyme, putative [Ricinus communis] |
| 9 |
Hb_005144_080 |
0.1207821566 |
- |
- |
Serine carboxypeptidase, putative [Ricinus communis] |
| 10 |
Hb_004282_010 |
0.1252784965 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At2g24230 [Jatropha curcas] |
| 11 |
Hb_027556_060 |
0.1270846465 |
- |
- |
hypothetical protein POPTR_0001s03520g [Populus trichocarpa] |
| 12 |
Hb_000714_060 |
0.1272091709 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 1-like [Populus euphratica] |
| 13 |
Hb_000011_370 |
0.1287628587 |
- |
- |
PREDICTED: uncharacterized protein LOC105631333 [Jatropha curcas] |
| 14 |
Hb_009767_050 |
0.1290730903 |
- |
- |
PREDICTED: uncharacterized protein LOC105631978 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000014_080 |
0.13252928 |
- |
- |
PREDICTED: UDP-glycosyltransferase 87A1-like [Jatropha curcas] |
| 16 |
Hb_002072_040 |
0.1350789314 |
- |
- |
PREDICTED: uncharacterized protein LOC105630881 [Jatropha curcas] |
| 17 |
Hb_007684_010 |
0.1355781521 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000749_120 |
0.1359276061 |
- |
- |
PREDICTED: uncharacterized protein LOC105644255 isoform X2 [Jatropha curcas] |
| 19 |
Hb_003754_020 |
0.1384688392 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 20 |
Hb_001473_060 |
0.1412965386 |
- |
- |
PREDICTED: gibberellin 2-beta-dioxygenase 8 [Jatropha curcas] |