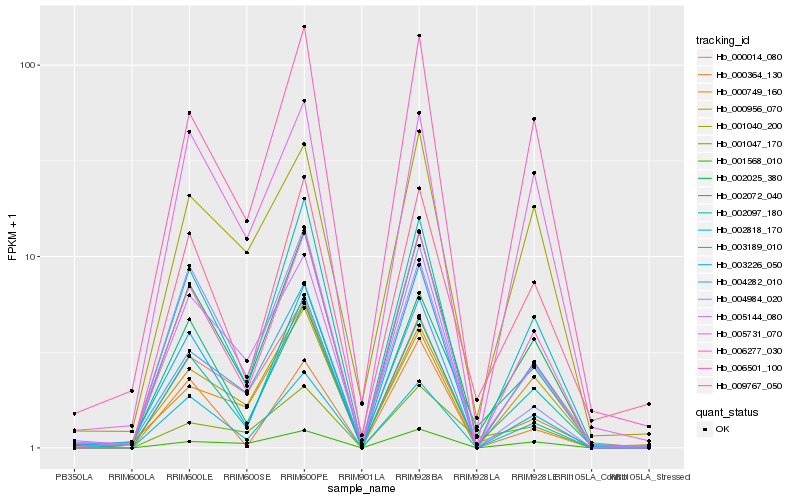
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002025_380 |
0.0 |
- |
- |
Uncharacterized protein TCM_011176 [Theobroma cacao] |
| 2 |
Hb_006277_030 |
0.0655903748 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_003189_010 |
0.0773942588 |
- |
- |
hypothetical protein EUGRSUZ_G02605 [Eucalyptus grandis] |
| 4 |
Hb_002818_170 |
0.0825849023 |
- |
- |
hypothetical protein POPTR_0014s10650g [Populus trichocarpa] |
| 5 |
Hb_002097_180 |
0.0987514944 |
- |
- |
PREDICTED: probable pectate lyase 12 isoform X1 [Jatropha curcas] |
| 6 |
Hb_005144_080 |
0.1011225096 |
- |
- |
Serine carboxypeptidase, putative [Ricinus communis] |
| 7 |
Hb_006501_100 |
0.1024816533 |
- |
- |
malic enzyme, putative [Ricinus communis] |
| 8 |
Hb_009767_050 |
0.1161088844 |
- |
- |
PREDICTED: uncharacterized protein LOC105631978 isoform X1 [Jatropha curcas] |
| 9 |
Hb_004282_010 |
0.1166564474 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At2g24230 [Jatropha curcas] |
| 10 |
Hb_002072_040 |
0.1170100804 |
- |
- |
PREDICTED: uncharacterized protein LOC105630881 [Jatropha curcas] |
| 11 |
Hb_001568_010 |
0.1323755624 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_005731_070 |
0.1331233275 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000014_080 |
0.1360813991 |
- |
- |
PREDICTED: UDP-glycosyltransferase 87A1-like [Jatropha curcas] |
| 14 |
Hb_001040_200 |
0.140804004 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA1H1-like [Jatropha curcas] |
| 15 |
Hb_000956_070 |
0.1428968676 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g01540 [Jatropha curcas] |
| 16 |
Hb_003226_050 |
0.1481773405 |
- |
- |
Pectinesterase-3 precursor, putative [Ricinus communis] |
| 17 |
Hb_004984_020 |
0.1493623059 |
- |
- |
PREDICTED: uncharacterized protein LOC105647814 [Jatropha curcas] |
| 18 |
Hb_001047_170 |
0.1521590443 |
- |
- |
PREDICTED: uncharacterized protein LOC105649981 [Jatropha curcas] |
| 19 |
Hb_000749_160 |
0.1527680996 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 20 |
Hb_000364_130 |
0.1539440628 |
- |
- |
unnamed protein product [Vitis vinifera] |