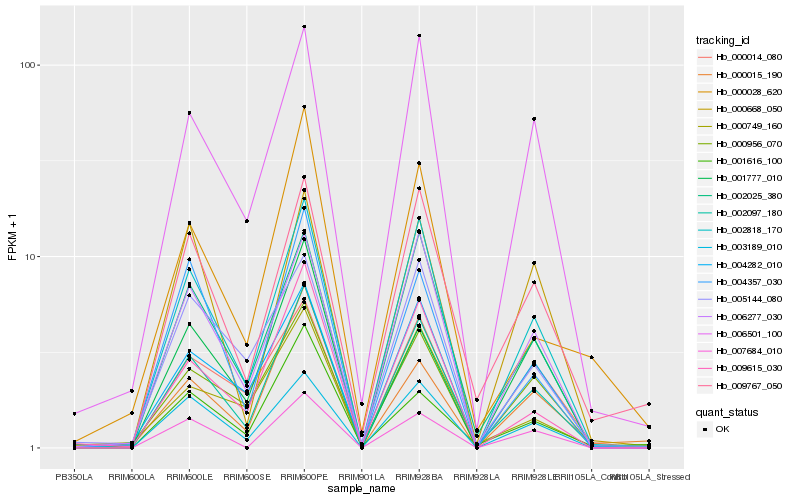
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002097_180 |
0.0 |
- |
- |
PREDICTED: probable pectate lyase 12 isoform X1 [Jatropha curcas] |
| 2 |
Hb_002818_170 |
0.0722530959 |
- |
- |
hypothetical protein POPTR_0014s10650g [Populus trichocarpa] |
| 3 |
Hb_002025_380 |
0.0987514944 |
- |
- |
Uncharacterized protein TCM_011176 [Theobroma cacao] |
| 4 |
Hb_003189_010 |
0.1039291197 |
- |
- |
hypothetical protein EUGRSUZ_G02605 [Eucalyptus grandis] |
| 5 |
Hb_006501_100 |
0.1146946891 |
- |
- |
malic enzyme, putative [Ricinus communis] |
| 6 |
Hb_006277_030 |
0.1160923872 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_009615_030 |
0.1210255202 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 8 |
Hb_009767_050 |
0.1235378137 |
- |
- |
PREDICTED: uncharacterized protein LOC105631978 isoform X1 [Jatropha curcas] |
| 9 |
Hb_004282_010 |
0.1274106399 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At2g24230 [Jatropha curcas] |
| 10 |
Hb_000956_070 |
0.1288942135 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g01540 [Jatropha curcas] |
| 11 |
Hb_007684_010 |
0.1343105642 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_004357_030 |
0.1396526988 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 13 |
Hb_000014_080 |
0.1435527529 |
- |
- |
PREDICTED: UDP-glycosyltransferase 87A1-like [Jatropha curcas] |
| 14 |
Hb_000668_050 |
0.1492065571 |
- |
- |
phosphoglycerate mutase, putative [Ricinus communis] |
| 15 |
Hb_005144_080 |
0.150131594 |
- |
- |
Serine carboxypeptidase, putative [Ricinus communis] |
| 16 |
Hb_000015_190 |
0.1522084174 |
- |
- |
PREDICTED: uncharacterized protein LOC105647810 [Jatropha curcas] |
| 17 |
Hb_001616_100 |
0.1529626936 |
- |
- |
PREDICTED: cell division control protein 45 homolog [Jatropha curcas] |
| 18 |
Hb_000028_620 |
0.1534265664 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000749_160 |
0.1534888266 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 20 |
Hb_001777_010 |
0.1546226212 |
- |
- |
- |