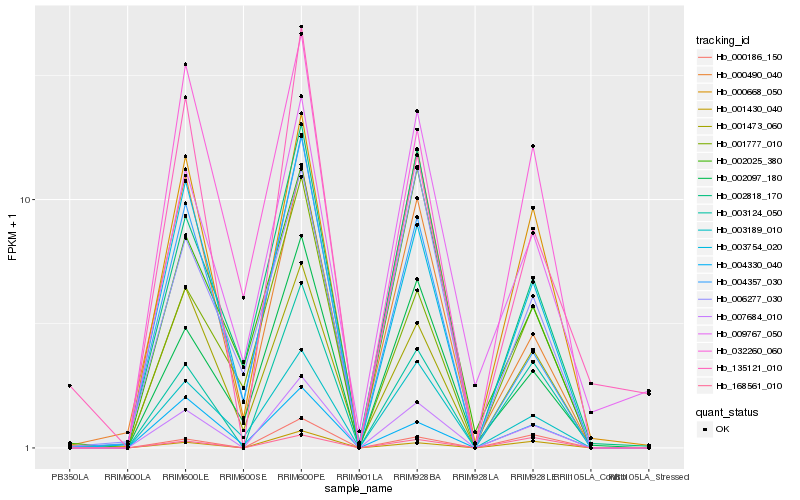
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007684_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_003124_050 |
0.0889448002 |
- |
- |
PREDICTED: uncharacterized protein LOC105641435 [Jatropha curcas] |
| 3 |
Hb_000668_050 |
0.1010713475 |
- |
- |
phosphoglycerate mutase, putative [Ricinus communis] |
| 4 |
Hb_003754_020 |
0.1292807125 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 5 |
Hb_002097_180 |
0.1343105642 |
- |
- |
PREDICTED: probable pectate lyase 12 isoform X1 [Jatropha curcas] |
| 6 |
Hb_003189_010 |
0.1355781521 |
- |
- |
hypothetical protein EUGRSUZ_G02605 [Eucalyptus grandis] |
| 7 |
Hb_002818_170 |
0.1419121806 |
- |
- |
hypothetical protein POPTR_0014s10650g [Populus trichocarpa] |
| 8 |
Hb_001473_060 |
0.1422937003 |
- |
- |
PREDICTED: gibberellin 2-beta-dioxygenase 8 [Jatropha curcas] |
| 9 |
Hb_001430_040 |
0.1426619449 |
- |
- |
gamma-tocopherol methyltransferase [Hevea brasiliensis] |
| 10 |
Hb_000186_150 |
0.1452792632 |
- |
- |
PREDICTED: light-induced protein, chloroplastic-like [Nicotiana sylvestris] |
| 11 |
Hb_004357_030 |
0.1539983282 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 12 |
Hb_004330_040 |
0.168525025 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000490_040 |
0.1687742878 |
- |
- |
PREDICTED: cyclin-D3-1 [Jatropha curcas] |
| 14 |
Hb_006277_030 |
0.1689763146 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_135121_010 |
0.1693209824 |
- |
- |
PREDICTED: uncharacterized protein LOC105635544 [Jatropha curcas] |
| 16 |
Hb_032260_060 |
0.1698697238 |
transcription factor |
TF Family: MYB-related |
Homeodomain-like superfamily protein [Theobroma cacao] |
| 17 |
Hb_001777_010 |
0.1725112976 |
- |
- |
- |
| 18 |
Hb_009767_050 |
0.1746568669 |
- |
- |
PREDICTED: uncharacterized protein LOC105631978 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002025_380 |
0.1761219215 |
- |
- |
Uncharacterized protein TCM_011176 [Theobroma cacao] |
| 20 |
Hb_168561_010 |
0.1766794766 |
- |
- |
PREDICTED: notchless protein homolog [Jatropha curcas] |