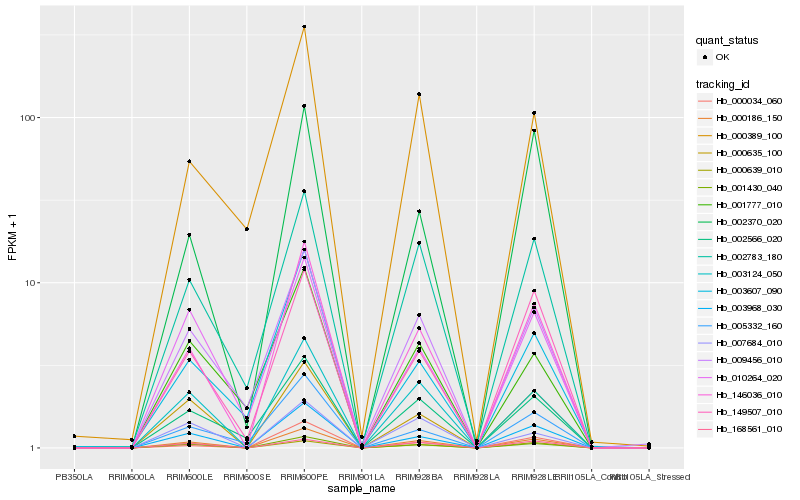
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000186_150 |
0.0 |
- |
- |
PREDICTED: light-induced protein, chloroplastic-like [Nicotiana sylvestris] |
| 2 |
Hb_001430_040 |
0.0410685524 |
- |
- |
gamma-tocopherol methyltransferase [Hevea brasiliensis] |
| 3 |
Hb_003124_050 |
0.0571066576 |
- |
- |
PREDICTED: uncharacterized protein LOC105641435 [Jatropha curcas] |
| 4 |
Hb_146036_010 |
0.063637171 |
- |
- |
hypothetical protein B456_005G052500 [Gossypium raimondii] |
| 5 |
Hb_003968_030 |
0.0799601273 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 6 |
Hb_000635_100 |
0.0898973147 |
- |
- |
- |
| 7 |
Hb_002783_180 |
0.1097904366 |
- |
- |
Tubulin beta-6 chain [Gossypium arboreum] |
| 8 |
Hb_009456_010 |
0.1162550583 |
- |
- |
PREDICTED: cinnamoyl-CoA reductase 1-like isoform X3 [Populus euphratica] |
| 9 |
Hb_002566_020 |
0.1171308436 |
- |
- |
hypothetical protein JCGZ_15085 [Jatropha curcas] |
| 10 |
Hb_149507_010 |
0.1306109768 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK9 [Malus domestica] |
| 11 |
Hb_168561_010 |
0.1325544137 |
- |
- |
PREDICTED: notchless protein homolog [Jatropha curcas] |
| 12 |
Hb_000639_010 |
0.1330565623 |
transcription factor |
TF Family: ERF |
hypothetical protein JCGZ_09727 [Jatropha curcas] |
| 13 |
Hb_010264_020 |
0.1381706449 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 14 |
Hb_007684_010 |
0.1452792632 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_005332_160 |
0.1464575207 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 2-like [Jatropha curcas] |
| 16 |
Hb_002370_020 |
0.1549542817 |
- |
- |
hypothetical protein JCGZ_19971 [Jatropha curcas] |
| 17 |
Hb_000389_100 |
0.156206444 |
- |
- |
phenylalanine ammonia-lyase 1 [Manihot esculenta] |
| 18 |
Hb_001777_010 |
0.1585761685 |
- |
- |
- |
| 19 |
Hb_000034_060 |
0.162356895 |
- |
- |
PREDICTED: O-acyltransferase WSD1-like [Citrus sinensis] |
| 20 |
Hb_003607_090 |
0.1656405706 |
- |
- |
PREDICTED: probable pectate lyase 8 [Jatropha curcas] |