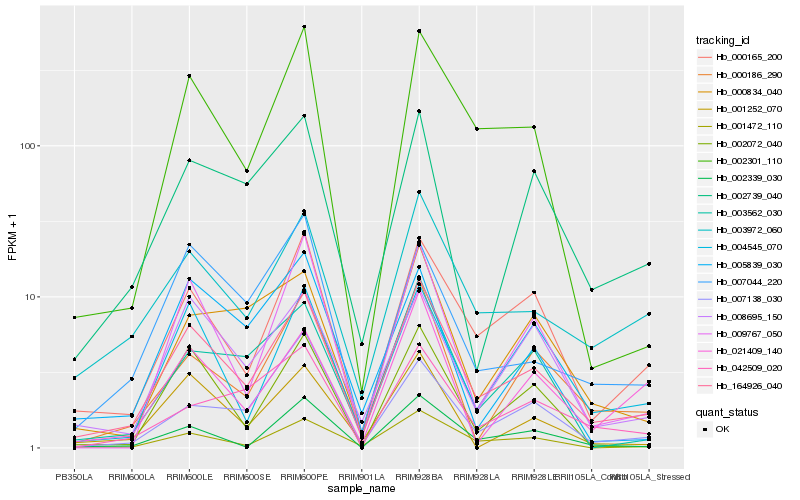
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_164926_040 |
0.0 |
- |
- |
Beta-1,3-galactosyltransferase sqv-2, putative [Ricinus communis] |
| 2 |
Hb_003972_060 |
0.1202364093 |
- |
- |
PREDICTED: UDP-arabinopyranose mutase 3 [Populus euphratica] |
| 3 |
Hb_000165_200 |
0.1268739241 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 4 |
Hb_001472_110 |
0.1352554381 |
desease resistance |
Gene Name: TIR_2 |
nucleoside-triphosphatase, putative [Ricinus communis] |
| 5 |
Hb_002301_110 |
0.1443342472 |
- |
- |
PREDICTED: protein E6-like [Populus euphratica] |
| 6 |
Hb_000186_290 |
0.147897465 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 8 isoform X1 [Jatropha curcas] |
| 7 |
Hb_008695_150 |
0.1553663805 |
- |
- |
PREDICTED: beta-1,3-galactosyltransferase 7 isoform X1 [Jatropha curcas] |
| 8 |
Hb_002739_040 |
0.156526977 |
- |
- |
PREDICTED: isocitrate dehydrogenase [NADP] [Jatropha curcas] |
| 9 |
Hb_004545_070 |
0.1586194345 |
- |
- |
PREDICTED: bifunctional 3-dehydroquinate dehydratase/shikimate dehydrogenase, chloroplastic-like [Jatropha curcas] |
| 10 |
Hb_009767_050 |
0.1594570433 |
- |
- |
PREDICTED: uncharacterized protein LOC105631978 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001252_070 |
0.159871466 |
- |
- |
hypothetical protein JCGZ_01385 [Jatropha curcas] |
| 12 |
Hb_002072_040 |
0.1613698572 |
- |
- |
PREDICTED: uncharacterized protein LOC105630881 [Jatropha curcas] |
| 13 |
Hb_003562_030 |
0.1624038027 |
- |
- |
- |
| 14 |
Hb_002339_030 |
0.1663888011 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 15 |
Hb_007044_220 |
0.1684035959 |
- |
- |
Allene oxide cyclase 4, chloroplast precursor, putative [Ricinus communis] |
| 16 |
Hb_000834_040 |
0.1695905392 |
- |
- |
PREDICTED: ras-related protein Rab7 [Cucumis melo] |
| 17 |
Hb_021409_140 |
0.1751332414 |
- |
- |
PREDICTED: trehalase isoform X1 [Jatropha curcas] |
| 18 |
Hb_042509_020 |
0.1762791855 |
- |
- |
hypothetical protein JCGZ_19555 [Jatropha curcas] |
| 19 |
Hb_007138_030 |
0.1822237334 |
- |
- |
PREDICTED: putative disease resistance protein At4g11170 [Jatropha curcas] |
| 20 |
Hb_005839_030 |
0.1837845273 |
- |
- |
PREDICTED: potassium transporter 2 [Jatropha curcas] |