| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
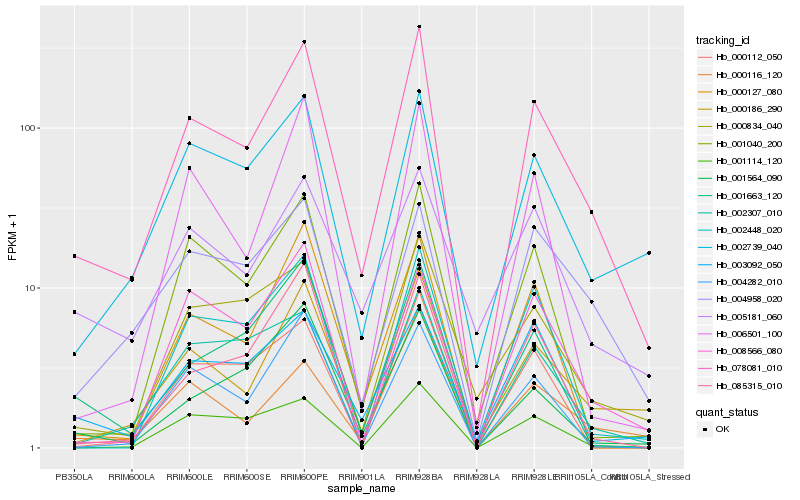
Hb_078081_010 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppa015726mg [Prunus persica] |
| 2 |
Hb_000186_290 |
0.1165124085 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 8 isoform X1 [Jatropha curcas] |
| 3 |
Hb_002739_040 |
0.1444903877 |
- |
- |
PREDICTED: isocitrate dehydrogenase [NADP] [Jatropha curcas] |
| 4 |
Hb_001040_200 |
0.1461662928 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA1H1-like [Jatropha curcas] |
| 5 |
Hb_000834_040 |
0.1509201088 |
- |
- |
PREDICTED: ras-related protein Rab7 [Cucumis melo] |
| 6 |
Hb_002307_010 |
0.1533393403 |
- |
- |
PREDICTED: uncharacterized protein LOC105632092 [Jatropha curcas] |
| 7 |
Hb_001663_120 |
0.1538943275 |
- |
- |
PREDICTED: formin-like protein 1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000116_120 |
0.154298332 |
- |
- |
PREDICTED: disease resistance response protein 206-like [Jatropha curcas] |
| 9 |
Hb_000127_080 |
0.1576168614 |
- |
- |
hypothetical protein POPTR_0005s21760g [Populus trichocarpa] |
| 10 |
Hb_006501_100 |
0.1610876445 |
- |
- |
malic enzyme, putative [Ricinus communis] |
| 11 |
Hb_004958_020 |
0.1612657761 |
- |
- |
PREDICTED: uncharacterized protein LOC104609039 [Nelumbo nucifera] |
| 12 |
Hb_001564_090 |
0.1614935086 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_008566_080 |
0.1636595388 |
- |
- |
PREDICTED: uncharacterized protein LOC105647834 [Jatropha curcas] |
| 14 |
Hb_085315_010 |
0.1642464098 |
- |
- |
PREDICTED: uncharacterized protein LOC105632092 [Jatropha curcas] |
| 15 |
Hb_003092_050 |
0.1653935184 |
- |
- |
PREDICTED: putative chloride channel-like protein CLC-g [Jatropha curcas] |
| 16 |
Hb_004282_010 |
0.1657813759 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At2g24230 [Jatropha curcas] |
| 17 |
Hb_005181_060 |
0.166440496 |
- |
- |
ATP-citrate synthase, putative [Ricinus communis] |
| 18 |
Hb_000112_050 |
0.1672090101 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL1 [Jatropha curcas] |
| 19 |
Hb_002448_020 |
0.1674643138 |
transcription factor |
TF Family: SBP |
Squamosa promoter-binding protein, putative [Ricinus communis] |
| 20 |
Hb_001114_120 |
0.1698148906 |
- |
- |
protein with unknown function [Ricinus communis] |