| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
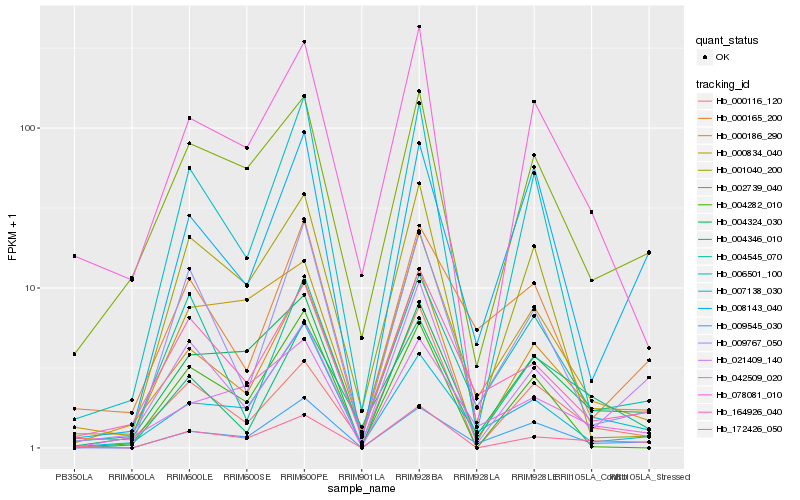
Hb_000186_290 |
0.0 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 8 isoform X1 [Jatropha curcas] |
| 2 |
Hb_078081_010 |
0.1165124085 |
- |
- |
hypothetical protein PRUPE_ppa015726mg [Prunus persica] |
| 3 |
Hb_002739_040 |
0.1359701786 |
- |
- |
PREDICTED: isocitrate dehydrogenase [NADP] [Jatropha curcas] |
| 4 |
Hb_164926_040 |
0.147897465 |
- |
- |
Beta-1,3-galactosyltransferase sqv-2, putative [Ricinus communis] |
| 5 |
Hb_000116_120 |
0.1514111332 |
- |
- |
PREDICTED: disease resistance response protein 206-like [Jatropha curcas] |
| 6 |
Hb_004346_010 |
0.1516643348 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_021409_140 |
0.1577014419 |
- |
- |
PREDICTED: trehalase isoform X1 [Jatropha curcas] |
| 8 |
Hb_172426_050 |
0.1578164187 |
- |
- |
PREDICTED: uncharacterized protein LOC105634266 [Jatropha curcas] |
| 9 |
Hb_004324_030 |
0.1626323566 |
- |
- |
PREDICTED: probable protein phosphatase 2C 8 [Jatropha curcas] |
| 10 |
Hb_006501_100 |
0.166254755 |
- |
- |
malic enzyme, putative [Ricinus communis] |
| 11 |
Hb_009545_030 |
0.1695313056 |
- |
- |
PREDICTED: folate-biopterin transporter 1, chloroplastic [Jatropha curcas] |
| 12 |
Hb_009767_050 |
0.1756480666 |
- |
- |
PREDICTED: uncharacterized protein LOC105631978 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001040_200 |
0.1762257327 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA1H1-like [Jatropha curcas] |
| 14 |
Hb_042509_020 |
0.1784658636 |
- |
- |
hypothetical protein JCGZ_19555 [Jatropha curcas] |
| 15 |
Hb_007138_030 |
0.1801074244 |
- |
- |
PREDICTED: putative disease resistance protein At4g11170 [Jatropha curcas] |
| 16 |
Hb_000834_040 |
0.1833624945 |
- |
- |
PREDICTED: ras-related protein Rab7 [Cucumis melo] |
| 17 |
Hb_004545_070 |
0.1833656711 |
- |
- |
PREDICTED: bifunctional 3-dehydroquinate dehydratase/shikimate dehydrogenase, chloroplastic-like [Jatropha curcas] |
| 18 |
Hb_004282_010 |
0.1848051903 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At2g24230 [Jatropha curcas] |
| 19 |
Hb_000165_200 |
0.1850253766 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 20 |
Hb_008143_040 |
0.1851511857 |
- |
- |
Alpha-expansin 4 precursor, putative [Ricinus communis] |