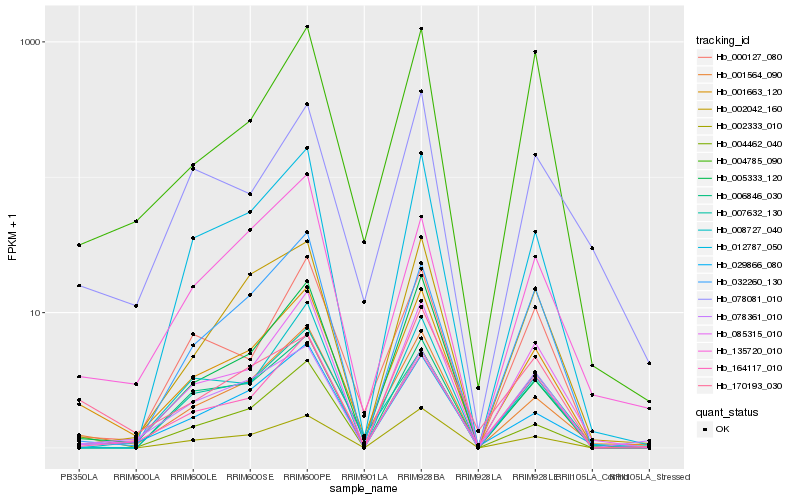
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001663_120 |
0.0 |
- |
- |
PREDICTED: formin-like protein 1 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001564_090 |
0.0869622495 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_029866_080 |
0.1242501651 |
- |
- |
PREDICTED: probable inactive receptor kinase At4g23740 [Jatropha curcas] |
| 4 |
Hb_085315_010 |
0.1327351949 |
- |
- |
PREDICTED: uncharacterized protein LOC105632092 [Jatropha curcas] |
| 5 |
Hb_004462_040 |
0.1357258681 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 6 |
Hb_164117_010 |
0.1413024996 |
- |
- |
hypothetical protein PHAVU_004G0480000g [Phaseolus vulgaris] |
| 7 |
Hb_005333_120 |
0.1426843636 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 2 [Jatropha curcas] |
| 8 |
Hb_012787_050 |
0.1456042506 |
- |
- |
PREDICTED: glutathione S-transferase U18-like [Jatropha curcas] |
| 9 |
Hb_000127_080 |
0.1506023049 |
- |
- |
hypothetical protein POPTR_0005s21760g [Populus trichocarpa] |
| 10 |
Hb_170193_030 |
0.1509382523 |
- |
- |
PREDICTED: probable disease resistance protein At5g63020 [Jatropha curcas] |
| 11 |
Hb_078081_010 |
0.1538943275 |
- |
- |
hypothetical protein PRUPE_ppa015726mg [Prunus persica] |
| 12 |
Hb_002333_010 |
0.1553340196 |
- |
- |
hypothetical protein JCGZ_18834 [Jatropha curcas] |
| 13 |
Hb_008727_040 |
0.1554423677 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002042_160 |
0.1558030801 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 15 |
Hb_006846_030 |
0.1561283638 |
- |
- |
respiratory burst oxidase A [Manihot esculenta] |
| 16 |
Hb_004785_090 |
0.156720511 |
- |
- |
hypothetical protein MIMGU_mgv1a018797mg [Erythranthe guttata] |
| 17 |
Hb_078361_010 |
0.1603082229 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 1 [Jatropha curcas] |
| 18 |
Hb_007632_130 |
0.1619555126 |
- |
- |
PREDICTED: nudix hydrolase 10-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_135720_010 |
0.1621956727 |
- |
- |
hypothetical protein JCGZ_17857 [Jatropha curcas] |
| 20 |
Hb_032260_130 |
0.1625856765 |
- |
- |
hypothetical protein POPTR_0003s13800g [Populus trichocarpa] |