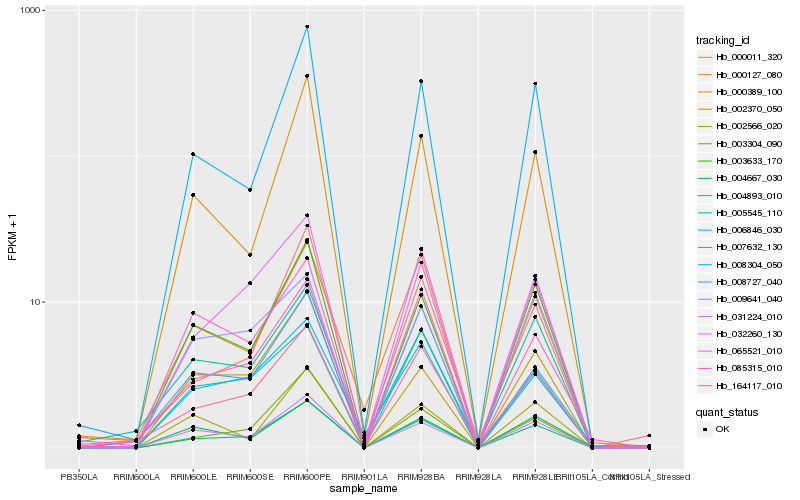
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004893_010 |
0.0 |
- |
- |
PREDICTED: O-acyltransferase WSD1-like [Jatropha curcas] |
| 2 |
Hb_008304_050 |
0.0874542742 |
- |
- |
PREDICTED: trans-cinnamate 4-monooxygenase [Jatropha curcas] |
| 3 |
Hb_003633_170 |
0.0884709403 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_032260_130 |
0.09319209 |
- |
- |
hypothetical protein POPTR_0003s13800g [Populus trichocarpa] |
| 5 |
Hb_031224_010 |
0.0949683349 |
- |
- |
- |
| 6 |
Hb_164117_010 |
0.0994005509 |
- |
- |
hypothetical protein PHAVU_004G0480000g [Phaseolus vulgaris] |
| 7 |
Hb_003304_090 |
0.1005710125 |
- |
- |
PREDICTED: uncharacterized protein LOC105633680 [Jatropha curcas] |
| 8 |
Hb_000389_100 |
0.1129001701 |
- |
- |
phenylalanine ammonia-lyase 1 [Manihot esculenta] |
| 9 |
Hb_085315_010 |
0.1165034675 |
- |
- |
PREDICTED: uncharacterized protein LOC105632092 [Jatropha curcas] |
| 10 |
Hb_000011_320 |
0.1235122378 |
- |
- |
PREDICTED: uncharacterized protein LOC105631328 [Jatropha curcas] |
| 11 |
Hb_007632_130 |
0.1263347598 |
- |
- |
PREDICTED: nudix hydrolase 10-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_005545_110 |
0.1331159609 |
- |
- |
PREDICTED: protein trichome birefringence-like 33 [Jatropha curcas] |
| 13 |
Hb_002370_050 |
0.1336105334 |
- |
- |
hypothetical protein JCGZ_18849 [Jatropha curcas] |
| 14 |
Hb_009641_040 |
0.1348397232 |
- |
- |
PREDICTED: uncharacterized protein LOC105633355 [Jatropha curcas] |
| 15 |
Hb_000127_080 |
0.1391963569 |
- |
- |
hypothetical protein POPTR_0005s21760g [Populus trichocarpa] |
| 16 |
Hb_006846_030 |
0.1398100132 |
- |
- |
respiratory burst oxidase A [Manihot esculenta] |
| 17 |
Hb_008727_040 |
0.1402754305 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_004667_030 |
0.141279658 |
- |
- |
hypothetical protein VITISV_034568 [Vitis vinifera] |
| 19 |
Hb_002566_020 |
0.1421197619 |
- |
- |
hypothetical protein JCGZ_15085 [Jatropha curcas] |
| 20 |
Hb_065521_010 |
0.1436502767 |
- |
- |
PREDICTED: probable metal-nicotianamine transporter YSL5 [Jatropha curcas] |