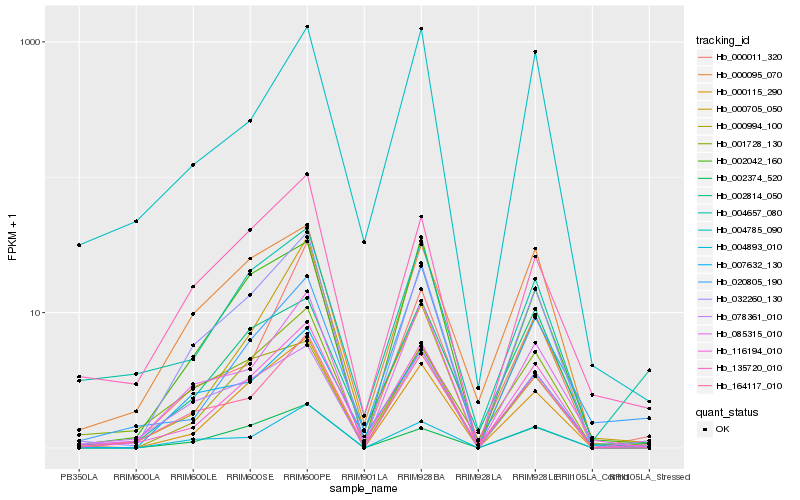
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_116194_010 |
0.0 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 2 |
Hb_032260_130 |
0.1011181404 |
- |
- |
hypothetical protein POPTR_0003s13800g [Populus trichocarpa] |
| 3 |
Hb_000115_290 |
0.1047452727 |
- |
- |
PREDICTED: uncharacterized protein LOC105124123 [Populus euphratica] |
| 4 |
Hb_164117_010 |
0.112403024 |
- |
- |
hypothetical protein PHAVU_004G0480000g [Phaseolus vulgaris] |
| 5 |
Hb_002042_160 |
0.129463647 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 6 |
Hb_002814_050 |
0.1322604203 |
- |
- |
PREDICTED: sugar carrier protein A [Jatropha curcas] |
| 7 |
Hb_004785_090 |
0.1368591605 |
- |
- |
hypothetical protein MIMGU_mgv1a018797mg [Erythranthe guttata] |
| 8 |
Hb_085315_010 |
0.1387886755 |
- |
- |
PREDICTED: uncharacterized protein LOC105632092 [Jatropha curcas] |
| 9 |
Hb_007632_130 |
0.1425009004 |
- |
- |
PREDICTED: nudix hydrolase 10-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_004893_010 |
0.1448806634 |
- |
- |
PREDICTED: O-acyltransferase WSD1-like [Jatropha curcas] |
| 11 |
Hb_000994_100 |
0.1482118892 |
- |
- |
RALFL33, putative [Ricinus communis] |
| 12 |
Hb_020805_190 |
0.1486869043 |
transcription factor |
TF Family: HB |
DNA binding protein, putative [Ricinus communis] |
| 13 |
Hb_078361_010 |
0.1514824722 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 1 [Jatropha curcas] |
| 14 |
Hb_000011_320 |
0.1528775362 |
- |
- |
PREDICTED: uncharacterized protein LOC105631328 [Jatropha curcas] |
| 15 |
Hb_004657_080 |
0.153044597 |
- |
- |
PREDICTED: probable inactive receptor kinase At5g67200 [Jatropha curcas] |
| 16 |
Hb_001728_130 |
0.1532144119 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 17 |
Hb_000705_050 |
0.1535202092 |
- |
- |
PREDICTED: uncharacterized protein C167.05 [Vitis vinifera] |
| 18 |
Hb_000095_070 |
0.1546717592 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_135720_010 |
0.1567836803 |
- |
- |
hypothetical protein JCGZ_17857 [Jatropha curcas] |
| 20 |
Hb_002374_520 |
0.1577055155 |
- |
- |
early nodulin 8 precursor family protein [Populus trichocarpa] |