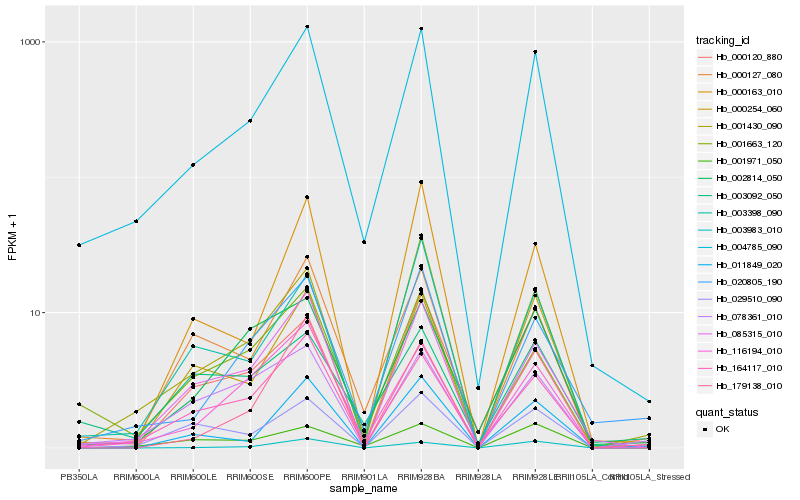
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004785_090 |
0.0 |
- |
- |
hypothetical protein MIMGU_mgv1a018797mg [Erythranthe guttata] |
| 2 |
Hb_164117_010 |
0.12523561 |
- |
- |
hypothetical protein PHAVU_004G0480000g [Phaseolus vulgaris] |
| 3 |
Hb_085315_010 |
0.135867169 |
- |
- |
PREDICTED: uncharacterized protein LOC105632092 [Jatropha curcas] |
| 4 |
Hb_116194_010 |
0.1368591605 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 5 |
Hb_001430_090 |
0.1429550661 |
- |
- |
hypothetical protein JCGZ_05131 [Jatropha curcas] |
| 6 |
Hb_000127_080 |
0.1454911971 |
- |
- |
hypothetical protein POPTR_0005s21760g [Populus trichocarpa] |
| 7 |
Hb_020805_190 |
0.148844827 |
transcription factor |
TF Family: HB |
DNA binding protein, putative [Ricinus communis] |
| 8 |
Hb_001971_050 |
0.1534700936 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 9 |
Hb_000254_060 |
0.1556849043 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001663_120 |
0.156720511 |
- |
- |
PREDICTED: formin-like protein 1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002814_050 |
0.1570284492 |
- |
- |
PREDICTED: sugar carrier protein A [Jatropha curcas] |
| 12 |
Hb_003398_090 |
0.1571281273 |
- |
- |
PREDICTED: expansin-A4 [Jatropha curcas] |
| 13 |
Hb_029510_090 |
0.1623885405 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At5g63020 [Jatropha curcas] |
| 14 |
Hb_003983_010 |
0.1679305127 |
- |
- |
serine/threonine-protein kinase bri1, putative [Ricinus communis] |
| 15 |
Hb_078361_010 |
0.171829264 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 1 [Jatropha curcas] |
| 16 |
Hb_000120_880 |
0.1729007403 |
- |
- |
PREDICTED: disease resistance response protein 206-like [Jatropha curcas] |
| 17 |
Hb_179138_010 |
0.1729061271 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 18 |
Hb_011849_020 |
0.1735725816 |
- |
- |
dioxygenase family protein [Populus trichocarpa] |
| 19 |
Hb_000163_010 |
0.1740216434 |
- |
- |
hypothetical protein JCGZ_13952 [Jatropha curcas] |
| 20 |
Hb_003092_050 |
0.1758273108 |
- |
- |
PREDICTED: putative chloride channel-like protein CLC-g [Jatropha curcas] |