| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
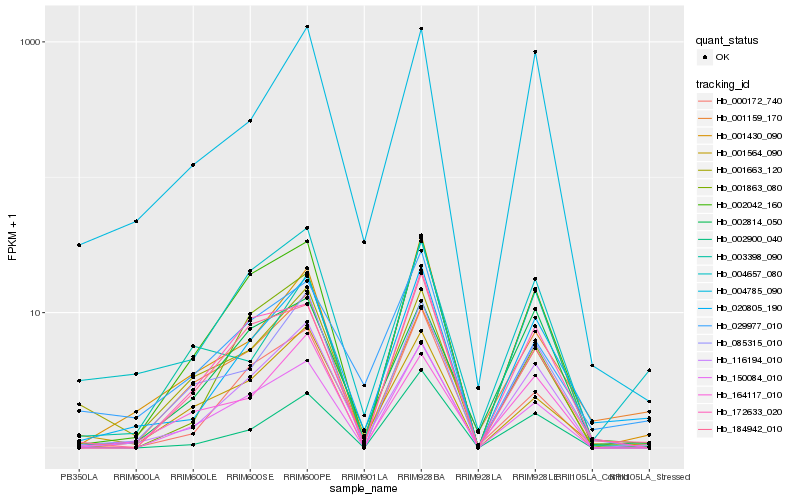
Hb_020805_190 |
0.0 |
transcription factor |
TF Family: HB |
DNA binding protein, putative [Ricinus communis] |
| 2 |
Hb_001430_090 |
0.1422635899 |
- |
- |
hypothetical protein JCGZ_05131 [Jatropha curcas] |
| 3 |
Hb_116194_010 |
0.1486869043 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 4 |
Hb_004785_090 |
0.148844827 |
- |
- |
hypothetical protein MIMGU_mgv1a018797mg [Erythranthe guttata] |
| 5 |
Hb_002042_160 |
0.1587963126 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 6 |
Hb_002900_040 |
0.1624199774 |
- |
- |
PREDICTED: 7-deoxyloganetic acid glucosyltransferase-like [Populus euphratica] |
| 7 |
Hb_085315_010 |
0.1637109176 |
- |
- |
PREDICTED: uncharacterized protein LOC105632092 [Jatropha curcas] |
| 8 |
Hb_029977_010 |
0.1661205631 |
- |
- |
catalytic, putative [Ricinus communis] |
| 9 |
Hb_184942_010 |
0.1682661018 |
- |
- |
STS14 protein precursor, putative [Ricinus communis] |
| 10 |
Hb_001159_170 |
0.1688237071 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 1 [Jatropha curcas] |
| 11 |
Hb_004657_080 |
0.1699215406 |
- |
- |
PREDICTED: probable inactive receptor kinase At5g67200 [Jatropha curcas] |
| 12 |
Hb_001564_090 |
0.1756841746 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_150084_010 |
0.1770567263 |
- |
- |
PREDICTED: premnaspirodiene oxygenase-like [Vitis vinifera] |
| 14 |
Hb_000172_740 |
0.1803505624 |
- |
- |
HXXXD-type acyl-transferase family protein, putative [Theobroma cacao] |
| 15 |
Hb_172633_020 |
0.1807322574 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g56140 [Populus euphratica] |
| 16 |
Hb_001663_120 |
0.1838578708 |
- |
- |
PREDICTED: formin-like protein 1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_002814_050 |
0.1845883102 |
- |
- |
PREDICTED: sugar carrier protein A [Jatropha curcas] |
| 18 |
Hb_001863_080 |
0.1901009875 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ATHB-6-like [Jatropha curcas] |
| 19 |
Hb_164117_010 |
0.1923497243 |
- |
- |
hypothetical protein PHAVU_004G0480000g [Phaseolus vulgaris] |
| 20 |
Hb_003398_090 |
0.1948346123 |
- |
- |
PREDICTED: expansin-A4 [Jatropha curcas] |