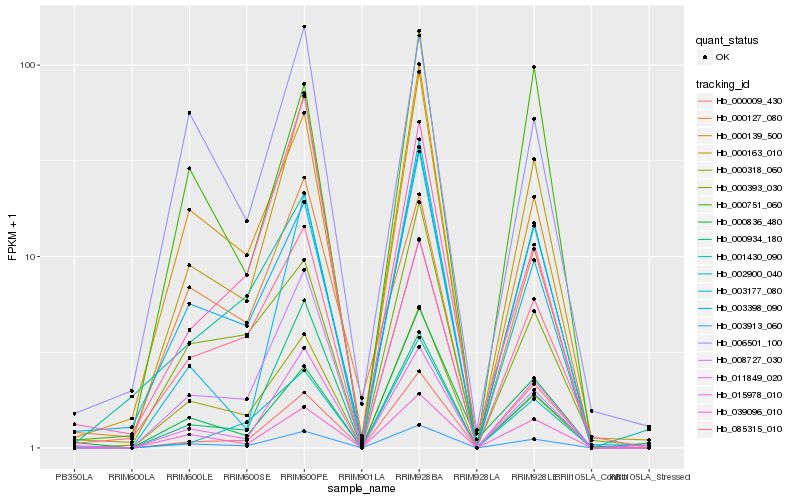
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000163_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_13952 [Jatropha curcas] |
| 2 |
Hb_003913_060 |
0.0754793905 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 3 |
Hb_011849_020 |
0.0856300635 |
- |
- |
dioxygenase family protein [Populus trichocarpa] |
| 4 |
Hb_003398_090 |
0.1157389973 |
- |
- |
PREDICTED: expansin-A4 [Jatropha curcas] |
| 5 |
Hb_015978_010 |
0.1191216831 |
- |
- |
PREDICTED: probable beta-D-xylosidase 2 [Populus euphratica] |
| 6 |
Hb_000318_060 |
0.1259520276 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_003177_080 |
0.1290426876 |
- |
- |
hypothetical protein L484_016032 [Morus notabilis] |
| 8 |
Hb_000139_500 |
0.1320524607 |
- |
- |
PREDICTED: putative uncharacterized protein DDB_G0286901 [Prunus mume] |
| 9 |
Hb_001430_090 |
0.1356089343 |
- |
- |
hypothetical protein JCGZ_05131 [Jatropha curcas] |
| 10 |
Hb_002900_040 |
0.1373762952 |
- |
- |
PREDICTED: 7-deoxyloganetic acid glucosyltransferase-like [Populus euphratica] |
| 11 |
Hb_085315_010 |
0.1548503913 |
- |
- |
PREDICTED: uncharacterized protein LOC105632092 [Jatropha curcas] |
| 12 |
Hb_000393_030 |
0.1600049762 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 13 |
Hb_008727_030 |
0.1613850882 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000934_180 |
0.161677187 |
- |
- |
hypothetical protein JCGZ_25237 [Jatropha curcas] |
| 15 |
Hb_000751_060 |
0.1650857324 |
- |
- |
PREDICTED: extensin-2-like, partial [Sesamum indicum] |
| 16 |
Hb_006501_100 |
0.1674892414 |
- |
- |
malic enzyme, putative [Ricinus communis] |
| 17 |
Hb_000009_430 |
0.1683905635 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000127_080 |
0.1685529406 |
- |
- |
hypothetical protein POPTR_0005s21760g [Populus trichocarpa] |
| 19 |
Hb_039096_010 |
0.1709686665 |
- |
- |
unknown [Populus trichocarpa] |
| 20 |
Hb_000836_480 |
0.1721112192 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |