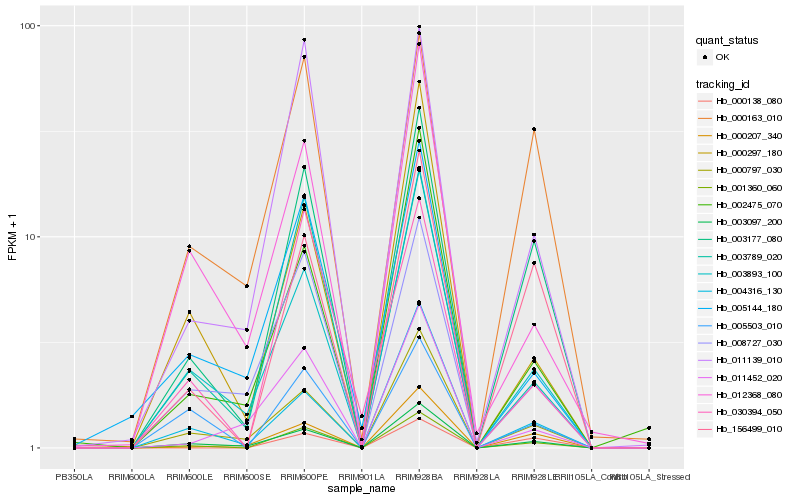
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001360_060 |
0.0 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 32 [Jatropha curcas] |
| 2 |
Hb_003177_080 |
0.0823601005 |
- |
- |
hypothetical protein L484_016032 [Morus notabilis] |
| 3 |
Hb_003789_020 |
0.1218603483 |
- |
- |
hypothetical protein JCGZ_23602 [Jatropha curcas] |
| 4 |
Hb_003097_200 |
0.1291540998 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_156499_010 |
0.1321092055 |
- |
- |
hypothetical protein JCGZ_21064 [Jatropha curcas] |
| 6 |
Hb_000797_030 |
0.1419133156 |
- |
- |
PREDICTED: leucoanthocyanidin dioxygenase [Jatropha curcas] |
| 7 |
Hb_011139_010 |
0.1432089602 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 8 |
Hb_003893_100 |
0.1462216834 |
- |
- |
PREDICTED: uncharacterized protein LOC105648774 [Jatropha curcas] |
| 9 |
Hb_005503_010 |
0.1485658046 |
- |
- |
PREDICTED: probable protein Pop3 [Jatropha curcas] |
| 10 |
Hb_030394_050 |
0.1511057929 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 11 |
Hb_008727_030 |
0.152487984 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000207_340 |
0.1584170384 |
transcription factor |
TF Family: GRAS |
hypothetical protein POPTR_0013s11930g [Populus trichocarpa] |
| 13 |
Hb_004316_130 |
0.1662277247 |
- |
- |
PREDICTED: glutathione transferase GST 23-like, partial [Eucalyptus grandis] |
| 14 |
Hb_005144_180 |
0.1662961466 |
- |
- |
PREDICTED: protein enabled homolog [Populus euphratica] |
| 15 |
Hb_012368_080 |
0.1755567748 |
- |
- |
PREDICTED: uncharacterized protein LOC105126277 [Populus euphratica] |
| 16 |
Hb_000138_080 |
0.177295582 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000163_010 |
0.1778924449 |
- |
- |
hypothetical protein JCGZ_13952 [Jatropha curcas] |
| 18 |
Hb_000297_180 |
0.1811943476 |
- |
- |
Desacetoxyvindoline 4-hydroxylase, putative [Ricinus communis] |
| 19 |
Hb_002475_070 |
0.1816680839 |
- |
- |
hypothetical protein POPTR_0017s02440g, partial [Populus trichocarpa] |
| 20 |
Hb_011452_020 |
0.1860585116 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 4 [Jatropha curcas] |