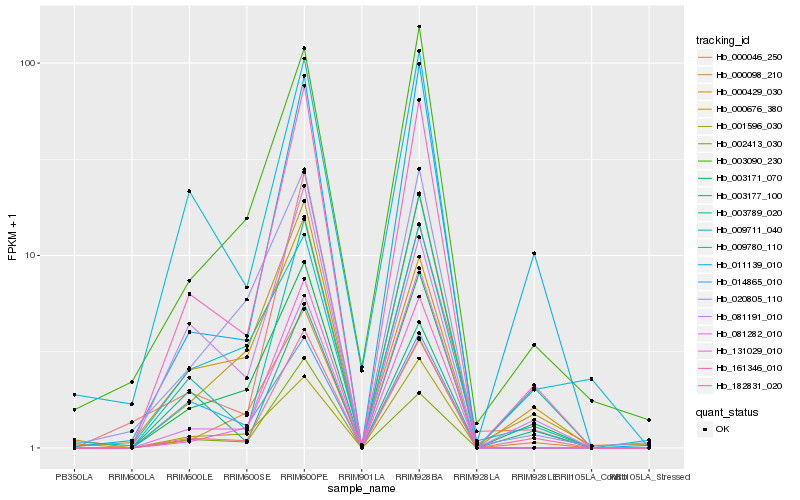
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_161346_010 |
0.0 |
- |
- |
PREDICTED: germin-like protein subfamily 1 member 20 [Populus euphratica] |
| 2 |
Hb_182831_020 |
0.063023301 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 3 |
Hb_003171_070 |
0.0999285414 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Citrus sinensis] |
| 4 |
Hb_131029_010 |
0.1121274924 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 5 |
Hb_000676_380 |
0.1210930676 |
- |
- |
cytochrome P450 family protein [Populus trichocarpa] |
| 6 |
Hb_002413_030 |
0.128595476 |
- |
- |
sulfate transporter, putative [Ricinus communis] |
| 7 |
Hb_003789_020 |
0.1299438314 |
- |
- |
hypothetical protein JCGZ_23602 [Jatropha curcas] |
| 8 |
Hb_081282_010 |
0.1358681707 |
- |
- |
PREDICTED: uncharacterized protein LOC105647834 [Jatropha curcas] |
| 9 |
Hb_000046_250 |
0.1366098033 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_003177_100 |
0.1394481332 |
- |
- |
PREDICTED: auxin-induced protein 6B [Jatropha curcas] |
| 11 |
Hb_011139_010 |
0.1398903425 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 12 |
Hb_009780_110 |
0.1399328323 |
- |
- |
PREDICTED: protein RSI-1 [Cicer arietinum] |
| 13 |
Hb_081191_010 |
0.1412128191 |
- |
- |
PREDICTED: pathogenesis-related protein PR-1-like [Jatropha curcas] |
| 14 |
Hb_001596_030 |
0.1432643728 |
- |
- |
PREDICTED: uncharacterized protein LOC105141672 [Populus euphratica] |
| 15 |
Hb_020805_110 |
0.145139179 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 12 [Jatropha curcas] |
| 16 |
Hb_003090_230 |
0.1490802517 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000098_210 |
0.1493167247 |
- |
- |
PREDICTED: uncharacterized protein LOC105633345 [Jatropha curcas] |
| 18 |
Hb_000429_030 |
0.1549063043 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 19 |
Hb_014865_010 |
0.1612636545 |
- |
- |
hypothetical protein PRUPE_ppa017366mg, partial [Prunus persica] |
| 20 |
Hb_009711_040 |
0.1617441257 |
- |
- |
PREDICTED: uncharacterized protein LOC105647789 [Jatropha curcas] |