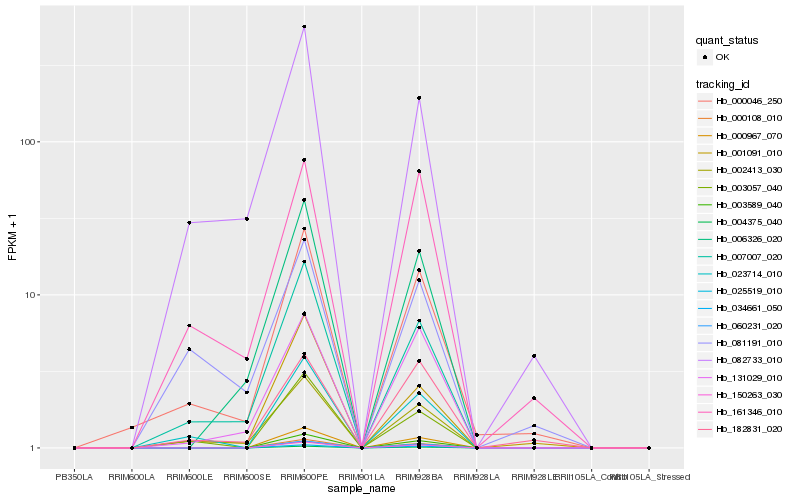
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000046_250 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002413_030 |
0.1124223082 |
- |
- |
sulfate transporter, putative [Ricinus communis] |
| 3 |
Hb_007007_020 |
0.1213659845 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_161346_010 |
0.1366098033 |
- |
- |
PREDICTED: germin-like protein subfamily 1 member 20 [Populus euphratica] |
| 5 |
Hb_023714_010 |
0.1369789248 |
- |
- |
hypothetical protein B456_007G377100 [Gossypium raimondii] |
| 6 |
Hb_082733_010 |
0.1388178624 |
- |
- |
hypothetical protein JCGZ_23600 [Jatropha curcas] |
| 7 |
Hb_182831_020 |
0.1457355852 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 8 |
Hb_131029_010 |
0.1476000873 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 9 |
Hb_003057_040 |
0.1488746081 |
- |
- |
PREDICTED: salt stress-induced hydrophobic peptide ESI3 [Vitis vinifera] |
| 10 |
Hb_006326_020 |
0.1569793026 |
- |
- |
- |
| 11 |
Hb_001091_010 |
0.1650273923 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 isoform X2 [Malus domestica] |
| 12 |
Hb_150263_030 |
0.1654368285 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000108_010 |
0.1655656241 |
- |
- |
hypothetical protein POPTR_0016s06140g, partial [Populus trichocarpa] |
| 14 |
Hb_004375_040 |
0.1655658383 |
- |
- |
hypothetical protein JCGZ_13965 [Jatropha curcas] |
| 15 |
Hb_003589_040 |
0.1656120466 |
- |
- |
hypothetical protein JCGZ_01204 [Jatropha curcas] |
| 16 |
Hb_060231_020 |
0.1659520199 |
- |
- |
PREDICTED: peroxisomal membrane protein PMP22 [Jatropha curcas] |
| 17 |
Hb_000967_070 |
0.1660997896 |
- |
- |
rubber peroxidase 1 [Hevea brasiliensis] |
| 18 |
Hb_025519_010 |
0.1661239282 |
- |
- |
PREDICTED: uncharacterized protein LOC103485710 [Cucumis melo] |
| 19 |
Hb_034661_050 |
0.1668266058 |
- |
- |
PREDICTED: zinc finger MYM-type protein 1-like, partial [Malus domestica] |
| 20 |
Hb_081191_010 |
0.1671919316 |
- |
- |
PREDICTED: pathogenesis-related protein PR-1-like [Jatropha curcas] |