| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
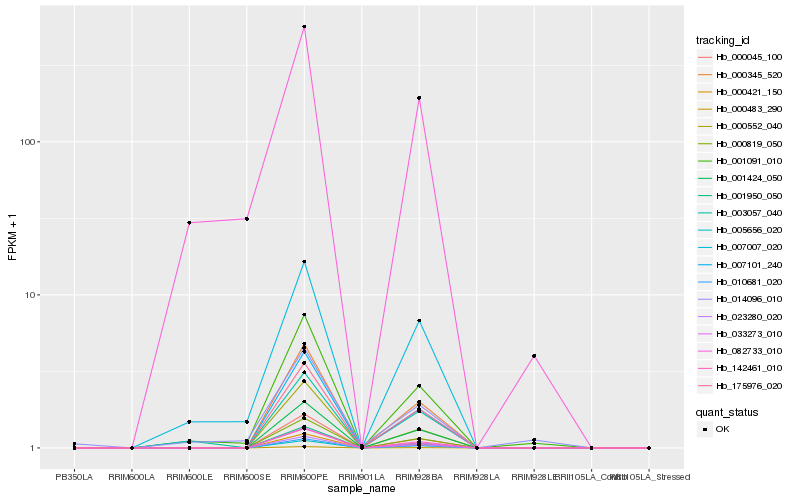
Hb_001091_010 |
0.0 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 isoform X2 [Malus domestica] |
| 2 |
Hb_007007_020 |
0.1148064158 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_082733_010 |
0.1234813667 |
- |
- |
hypothetical protein JCGZ_23600 [Jatropha curcas] |
| 4 |
Hb_000421_150 |
0.1245160084 |
- |
- |
Zinc finger protein GIS2 [Medicago truncatula] |
| 5 |
Hb_007101_240 |
0.1245255113 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_010681_020 |
0.1245424464 |
- |
- |
JHL22C18.10 [Jatropha curcas] |
| 7 |
Hb_000483_290 |
0.1245474607 |
- |
- |
PREDICTED: ankyrin-1-like [Prunus mume] |
| 8 |
Hb_000045_100 |
0.1251365212 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000345_520 |
0.125438462 |
- |
- |
hypothetical protein POPTR_0013s02440g [Populus trichocarpa] |
| 10 |
Hb_023280_020 |
0.1256089782 |
- |
- |
hypothetical protein JCGZ_21650 [Jatropha curcas] |
| 11 |
Hb_033273_010 |
0.1260342618 |
- |
- |
- |
| 12 |
Hb_000819_050 |
0.1260387084 |
- |
- |
hypothetical protein Csa_4G290160 [Cucumis sativus] |
| 13 |
Hb_142461_010 |
0.128797737 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 14 |
Hb_001950_050 |
0.1288365498 |
- |
- |
- |
| 15 |
Hb_014096_010 |
0.1302453934 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 16 |
Hb_000552_040 |
0.1307015219 |
- |
- |
- |
| 17 |
Hb_175976_020 |
0.130979508 |
- |
- |
PREDICTED: testis-expressed sequence 10 protein isoform X2 [Jatropha curcas] |
| 18 |
Hb_003057_040 |
0.1322941268 |
- |
- |
PREDICTED: salt stress-induced hydrophobic peptide ESI3 [Vitis vinifera] |
| 19 |
Hb_001424_050 |
0.1329297092 |
- |
- |
PREDICTED: zinc finger protein HD1-like isoform X2 [Jatropha curcas] |
| 20 |
Hb_005656_020 |
0.1334502208 |
- |
- |
PREDICTED: uncharacterized protein LOC102707106 [Oryza brachyantha] |