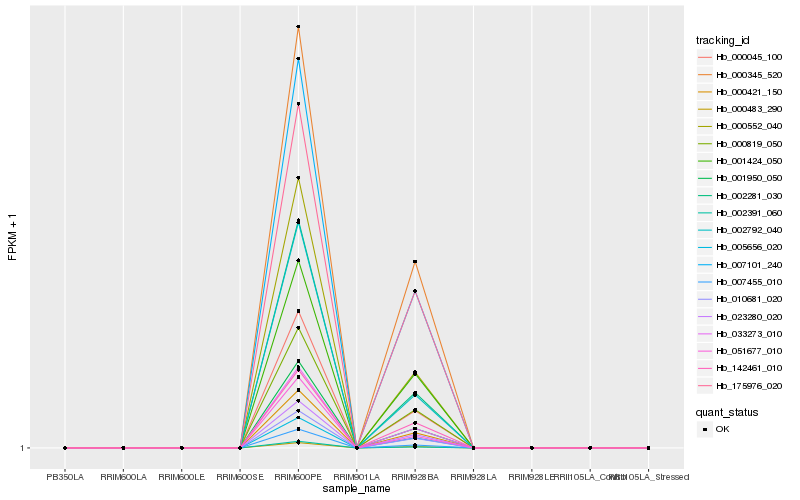
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000045_100 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_033273_010 |
0.00711062 |
- |
- |
- |
| 3 |
Hb_000421_150 |
0.0129811205 |
- |
- |
Zinc finger protein GIS2 [Medicago truncatula] |
| 4 |
Hb_007101_240 |
0.0141598868 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_010681_020 |
0.0151724751 |
- |
- |
JHL22C18.10 [Jatropha curcas] |
| 6 |
Hb_000483_290 |
0.0154041629 |
- |
- |
PREDICTED: ankyrin-1-like [Prunus mume] |
| 7 |
Hb_001950_050 |
0.0207989066 |
- |
- |
- |
| 8 |
Hb_000552_040 |
0.0274997304 |
- |
- |
- |
| 9 |
Hb_000345_520 |
0.0278582896 |
- |
- |
hypothetical protein POPTR_0013s02440g [Populus trichocarpa] |
| 10 |
Hb_023280_020 |
0.029217658 |
- |
- |
hypothetical protein JCGZ_21650 [Jatropha curcas] |
| 11 |
Hb_000819_050 |
0.0322378917 |
- |
- |
hypothetical protein Csa_4G290160 [Cucumis sativus] |
| 12 |
Hb_002281_030 |
0.0416987841 |
- |
- |
- |
| 13 |
Hb_142461_010 |
0.0457376495 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 14 |
Hb_002792_040 |
0.0460947042 |
- |
- |
- |
| 15 |
Hb_175976_020 |
0.0534980219 |
- |
- |
PREDICTED: testis-expressed sequence 10 protein isoform X2 [Jatropha curcas] |
| 16 |
Hb_001424_050 |
0.0594448685 |
- |
- |
PREDICTED: zinc finger protein HD1-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_005656_020 |
0.0609220303 |
- |
- |
PREDICTED: uncharacterized protein LOC102707106 [Oryza brachyantha] |
| 18 |
Hb_007455_010 |
0.0610427106 |
- |
- |
PREDICTED: uncharacterized protein LOC105800996 [Gossypium raimondii] |
| 19 |
Hb_051677_010 |
0.0646823349 |
- |
- |
Uncharacterized protein TCM_017953 [Theobroma cacao] |
| 20 |
Hb_002391_060 |
0.065476691 |
- |
- |
PREDICTED: uncharacterized protein LOC100853799 [Vitis vinifera] |