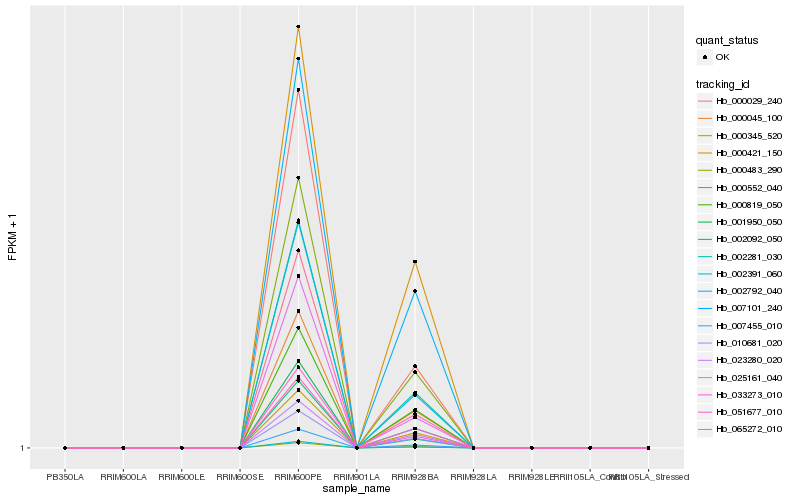
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000552_040 |
0.0 |
- |
- |
- |
| 2 |
Hb_001950_050 |
0.0067042426 |
- |
- |
- |
| 3 |
Hb_002281_030 |
0.01421419 |
- |
- |
- |
| 4 |
Hb_002792_040 |
0.0186170792 |
- |
- |
- |
| 5 |
Hb_033273_010 |
0.0203925733 |
- |
- |
- |
| 6 |
Hb_000045_100 |
0.0274997304 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_007455_010 |
0.0335974012 |
- |
- |
PREDICTED: uncharacterized protein LOC105800996 [Gossypium raimondii] |
| 8 |
Hb_051677_010 |
0.0372470017 |
- |
- |
Uncharacterized protein TCM_017953 [Theobroma cacao] |
| 9 |
Hb_002391_060 |
0.0380436491 |
- |
- |
PREDICTED: uncharacterized protein LOC100853799 [Vitis vinifera] |
| 10 |
Hb_000421_150 |
0.0404687729 |
- |
- |
Zinc finger protein GIS2 [Medicago truncatula] |
| 11 |
Hb_007101_240 |
0.041646089 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_010681_020 |
0.042657386 |
- |
- |
JHL22C18.10 [Jatropha curcas] |
| 13 |
Hb_000483_290 |
0.0428887725 |
- |
- |
PREDICTED: ankyrin-1-like [Prunus mume] |
| 14 |
Hb_025161_040 |
0.049484705 |
- |
- |
kinase, putative [Ricinus communis] |
| 15 |
Hb_000345_520 |
0.0553235536 |
- |
- |
hypothetical protein POPTR_0013s02440g [Populus trichocarpa] |
| 16 |
Hb_000029_240 |
0.0560863554 |
- |
- |
PREDICTED: pre-mRNA-splicing factor SLU7-A-like [Elaeis guineensis] |
| 17 |
Hb_023280_020 |
0.0566804464 |
- |
- |
hypothetical protein JCGZ_21650 [Jatropha curcas] |
| 18 |
Hb_000819_050 |
0.0596949321 |
- |
- |
hypothetical protein Csa_4G290160 [Cucumis sativus] |
| 19 |
Hb_065272_010 |
0.0612191431 |
- |
- |
hypothetical protein JCGZ_06349 [Jatropha curcas] |
| 20 |
Hb_002092_050 |
0.0654577629 |
- |
- |
hypothetical protein JCGZ_05877 [Jatropha curcas] |