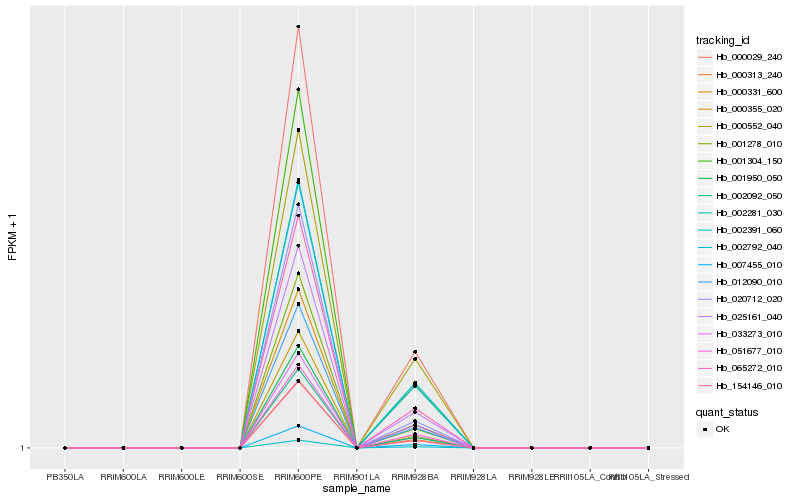
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_025161_040 |
0.0 |
- |
- |
kinase, putative [Ricinus communis] |
| 2 |
Hb_000029_240 |
0.0066235658 |
- |
- |
PREDICTED: pre-mRNA-splicing factor SLU7-A-like [Elaeis guineensis] |
| 3 |
Hb_002391_060 |
0.0114657827 |
- |
- |
PREDICTED: uncharacterized protein LOC100853799 [Vitis vinifera] |
| 4 |
Hb_065272_010 |
0.0117774625 |
- |
- |
hypothetical protein JCGZ_06349 [Jatropha curcas] |
| 5 |
Hb_051677_010 |
0.0122635515 |
- |
- |
Uncharacterized protein TCM_017953 [Theobroma cacao] |
| 6 |
Hb_007455_010 |
0.0159173252 |
- |
- |
PREDICTED: uncharacterized protein LOC105800996 [Gossypium raimondii] |
| 7 |
Hb_002092_050 |
0.0160362941 |
- |
- |
hypothetical protein JCGZ_05877 [Jatropha curcas] |
| 8 |
Hb_000331_600 |
0.0187391906 |
- |
- |
PREDICTED: probable Ufm1-specific protease [Jatropha curcas] |
| 9 |
Hb_012090_010 |
0.0215644098 |
- |
- |
hypothetical protein RCOM_0904330 [Ricinus communis] |
| 10 |
Hb_000313_240 |
0.0220944759 |
- |
- |
hypothetical protein RCOM_0377590 [Ricinus communis] |
| 11 |
Hb_002792_040 |
0.0308988857 |
- |
- |
- |
| 12 |
Hb_001304_150 |
0.0340871489 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 13 |
Hb_002281_030 |
0.0352975224 |
- |
- |
- |
| 14 |
Hb_154146_010 |
0.0369284251 |
- |
- |
hypothetical protein JCGZ_00346 [Jatropha curcas] |
| 15 |
Hb_000552_040 |
0.049484705 |
- |
- |
- |
| 16 |
Hb_001950_050 |
0.0561695599 |
- |
- |
- |
| 17 |
Hb_001278_010 |
0.0573517454 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000355_020 |
0.0597118461 |
- |
- |
- |
| 19 |
Hb_020712_020 |
0.0688844181 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ATX4 [Elaeis guineensis] |
| 20 |
Hb_033273_010 |
0.0698060622 |
- |
- |
- |