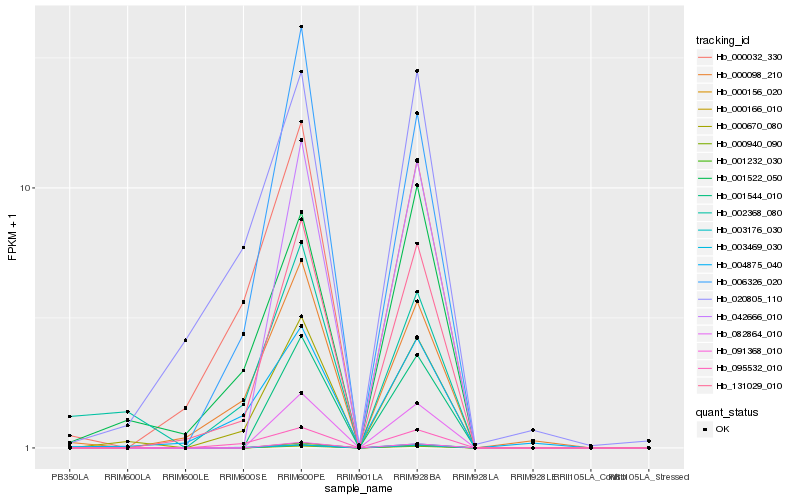
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000670_080 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_131029_010 |
0.1107549837 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 3 |
Hb_095532_010 |
0.1298590766 |
- |
- |
BnaC05g27570D [Brassica napus] |
| 4 |
Hb_001522_050 |
0.1358075692 |
- |
- |
PREDICTED: CLAVATA3/ESR (CLE)-related protein 25 [Jatropha curcas] |
| 5 |
Hb_006326_020 |
0.1366332316 |
- |
- |
- |
| 6 |
Hb_000032_330 |
0.1475916078 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_004875_040 |
0.154687158 |
- |
- |
PREDICTED: uncharacterized protein LOC105645797 [Jatropha curcas] |
| 8 |
Hb_000098_210 |
0.1598141249 |
- |
- |
PREDICTED: uncharacterized protein LOC105633345 [Jatropha curcas] |
| 9 |
Hb_003469_030 |
0.1613491483 |
- |
- |
hypothetical protein JCGZ_01307 [Jatropha curcas] |
| 10 |
Hb_002368_080 |
0.1614729227 |
- |
- |
PREDICTED: transcription factor SCREAM2-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_020805_110 |
0.1654404888 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 12 [Jatropha curcas] |
| 12 |
Hb_001544_010 |
0.166691645 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000166_010 |
0.1667503205 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |
| 14 |
Hb_082864_010 |
0.1667571468 |
- |
- |
JHL25H03.10 [Jatropha curcas] |
| 15 |
Hb_091368_010 |
0.1669057452 |
- |
- |
gibberellin 20-oxidase, putative [Ricinus communis] |
| 16 |
Hb_042666_010 |
0.1674966545 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 17 |
Hb_001232_030 |
0.1677550146 |
- |
- |
hypothetical protein POPTR_0001s18270g [Populus trichocarpa] |
| 18 |
Hb_000940_090 |
0.1684386251 |
- |
- |
hypothetical protein CISIN_1g046520mg [Citrus sinensis] |
| 19 |
Hb_000156_020 |
0.1687102029 |
- |
- |
PREDICTED: la-related protein 6A [Jatropha curcas] |
| 20 |
Hb_003176_030 |
0.1687102029 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Solanum lycopersicum] |