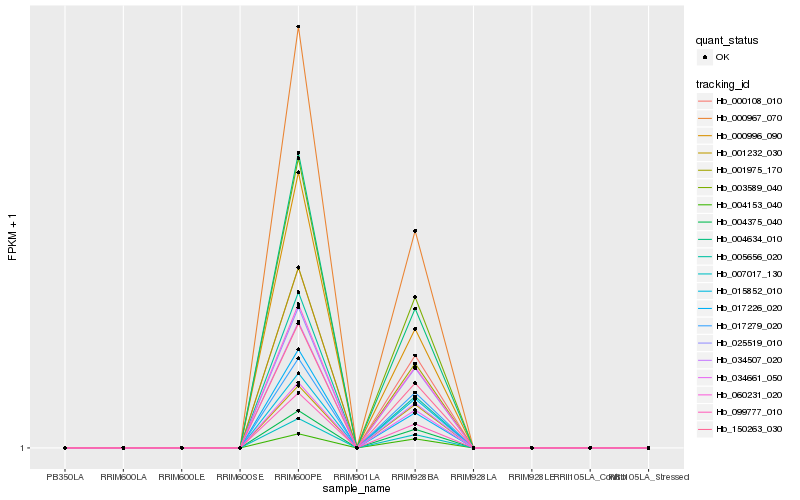
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000967_070 |
0.0 |
- |
- |
rubber peroxidase 1 [Hevea brasiliensis] |
| 2 |
Hb_003589_040 |
0.0071737735 |
- |
- |
hypothetical protein JCGZ_01204 [Jatropha curcas] |
| 3 |
Hb_004375_040 |
0.0082252342 |
- |
- |
hypothetical protein JCGZ_13965 [Jatropha curcas] |
| 4 |
Hb_000108_010 |
0.0082304767 |
- |
- |
hypothetical protein POPTR_0016s06140g, partial [Populus trichocarpa] |
| 5 |
Hb_001975_170 |
0.0108821445 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 26A [Nelumbo nucifera] |
| 6 |
Hb_007017_130 |
0.0115605879 |
- |
- |
PREDICTED: glycerol-3-phosphate 2-O-acyltransferase 6-like isoform X1 [Populus euphratica] |
| 7 |
Hb_150263_030 |
0.0130631922 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_004634_010 |
0.013503822 |
- |
- |
hypothetical protein POPTR_0002s23100g [Populus trichocarpa] |
| 9 |
Hb_099777_010 |
0.0198644853 |
- |
- |
PREDICTED: uncharacterized protein LOC104884611 [Beta vulgaris subsp. vulgaris] |
| 10 |
Hb_060231_020 |
0.0285986588 |
- |
- |
PREDICTED: peroxisomal membrane protein PMP22 [Jatropha curcas] |
| 11 |
Hb_025519_010 |
0.0306316703 |
- |
- |
PREDICTED: uncharacterized protein LOC103485710 [Cucumis melo] |
| 12 |
Hb_000996_090 |
0.0308503826 |
transcription factor |
TF Family: NF-YC |
ccaat-binding transcription factor, putative [Ricinus communis] |
| 13 |
Hb_034661_050 |
0.0370724658 |
- |
- |
PREDICTED: zinc finger MYM-type protein 1-like, partial [Malus domestica] |
| 14 |
Hb_017279_020 |
0.0498550233 |
- |
- |
protein MOTHER of FT and TF 1 [Jatropha curcas] |
| 15 |
Hb_004153_040 |
0.0598365181 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 16 |
Hb_017226_020 |
0.060711028 |
transcription factor |
TF Family: MYB |
hypothetical protein JCGZ_06720 [Jatropha curcas] |
| 17 |
Hb_034507_020 |
0.0618442931 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 18 |
Hb_015852_010 |
0.0632826197 |
- |
- |
PREDICTED: uncharacterized protein LOC104106381 [Nicotiana tomentosiformis] |
| 19 |
Hb_001232_030 |
0.0753957842 |
- |
- |
hypothetical protein POPTR_0001s18270g [Populus trichocarpa] |
| 20 |
Hb_005656_020 |
0.0772504602 |
- |
- |
PREDICTED: uncharacterized protein LOC102707106 [Oryza brachyantha] |