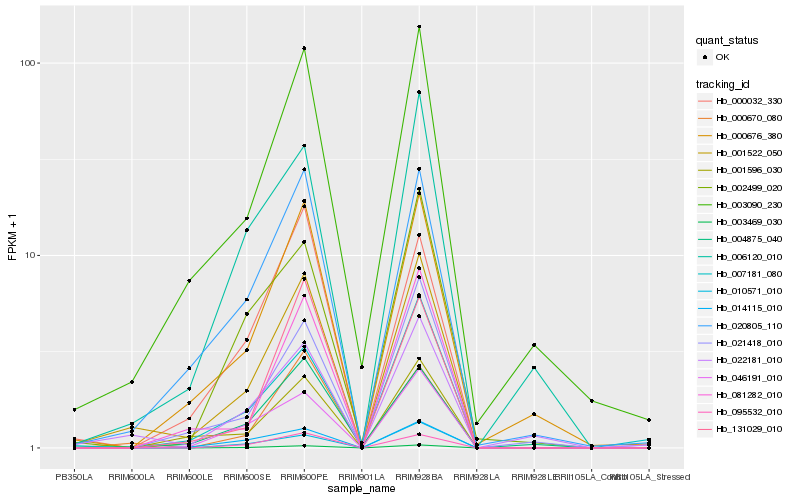
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001522_050 |
0.0 |
- |
- |
PREDICTED: CLAVATA3/ESR (CLE)-related protein 25 [Jatropha curcas] |
| 2 |
Hb_003469_030 |
0.119431849 |
- |
- |
hypothetical protein JCGZ_01307 [Jatropha curcas] |
| 3 |
Hb_020805_110 |
0.1208450309 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 12 [Jatropha curcas] |
| 4 |
Hb_003090_230 |
0.1218403486 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_022181_010 |
0.1293803051 |
- |
- |
PREDICTED: peroxidase 19 [Jatropha curcas] |
| 6 |
Hb_000676_380 |
0.1349062853 |
- |
- |
cytochrome P450 family protein [Populus trichocarpa] |
| 7 |
Hb_000670_080 |
0.1358075692 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001596_030 |
0.1373720033 |
- |
- |
PREDICTED: uncharacterized protein LOC105141672 [Populus euphratica] |
| 9 |
Hb_081282_010 |
0.1376001043 |
- |
- |
PREDICTED: uncharacterized protein LOC105647834 [Jatropha curcas] |
| 10 |
Hb_046191_010 |
0.1380972447 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 11 |
Hb_004875_040 |
0.1440254691 |
- |
- |
PREDICTED: uncharacterized protein LOC105645797 [Jatropha curcas] |
| 12 |
Hb_021418_010 |
0.148341004 |
- |
- |
PREDICTED: probable pectate lyase 12 [Prunus mume] |
| 13 |
Hb_006120_010 |
0.1496910361 |
- |
- |
invertase inhibitor [Manihot esculenta] |
| 14 |
Hb_095532_010 |
0.1526222832 |
- |
- |
BnaC05g27570D [Brassica napus] |
| 15 |
Hb_014115_010 |
0.1527997289 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 16 |
Hb_010571_010 |
0.1577984546 |
- |
- |
PREDICTED: non-symbiotic hemoglobin 2 [Jatropha curcas] |
| 17 |
Hb_002499_020 |
0.1614700509 |
- |
- |
hevamine-A precursor, putative [Ricinus communis] |
| 18 |
Hb_131029_010 |
0.1616588878 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 19 |
Hb_007181_080 |
0.1624818083 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 20 |
Hb_000032_330 |
0.163414591 |
- |
- |
conserved hypothetical protein [Ricinus communis] |