| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
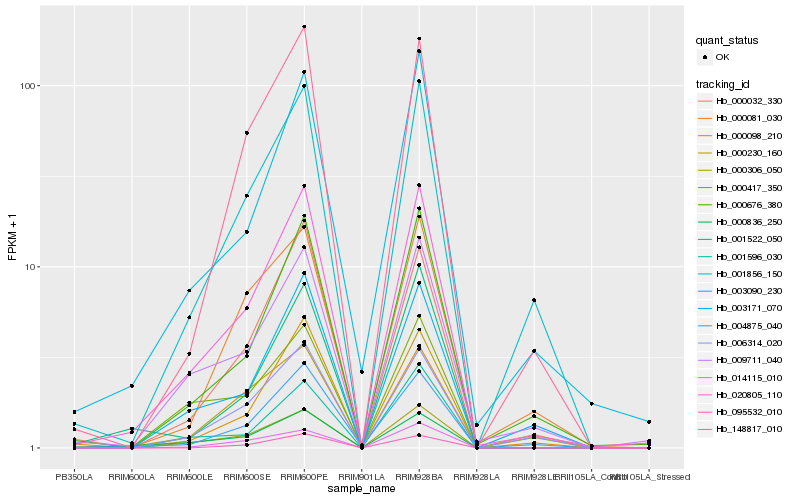
Hb_020805_110 |
0.0 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 12 [Jatropha curcas] |
| 2 |
Hb_000676_380 |
0.0873445868 |
- |
- |
cytochrome P450 family protein [Populus trichocarpa] |
| 3 |
Hb_009711_040 |
0.088901136 |
- |
- |
PREDICTED: uncharacterized protein LOC105647789 [Jatropha curcas] |
| 4 |
Hb_004875_040 |
0.0912600032 |
- |
- |
PREDICTED: uncharacterized protein LOC105645797 [Jatropha curcas] |
| 5 |
Hb_000306_050 |
0.0924565487 |
transcription factor |
TF Family: PLATZ |
protein with unknown function [Ricinus communis] |
| 6 |
Hb_003090_230 |
0.0972825237 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001856_150 |
0.1039453758 |
- |
- |
- |
| 8 |
Hb_003171_070 |
0.1050252656 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Citrus sinensis] |
| 9 |
Hb_000032_330 |
0.1105443472 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_148817_010 |
0.1127202187 |
- |
- |
PREDICTED: CASP-like protein 1D1 [Jatropha curcas] |
| 11 |
Hb_001596_030 |
0.1138866284 |
- |
- |
PREDICTED: uncharacterized protein LOC105141672 [Populus euphratica] |
| 12 |
Hb_000836_250 |
0.1147882051 |
- |
- |
hypothetical protein VITISV_022299 [Vitis vinifera] |
| 13 |
Hb_006314_020 |
0.1173351793 |
- |
- |
PREDICTED: GABA transporter 1-like [Jatropha curcas] |
| 14 |
Hb_001522_050 |
0.1208450309 |
- |
- |
PREDICTED: CLAVATA3/ESR (CLE)-related protein 25 [Jatropha curcas] |
| 15 |
Hb_014115_010 |
0.1262533347 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 16 |
Hb_000417_350 |
0.1275441303 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000098_210 |
0.1320250331 |
- |
- |
PREDICTED: uncharacterized protein LOC105633345 [Jatropha curcas] |
| 18 |
Hb_000230_160 |
0.1344016498 |
- |
- |
PREDICTED: uncharacterized protein LOC105631858 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000081_030 |
0.1367381748 |
- |
- |
BnaC09g05810D [Brassica napus] |
| 20 |
Hb_095532_010 |
0.1381078543 |
- |
- |
BnaC05g27570D [Brassica napus] |