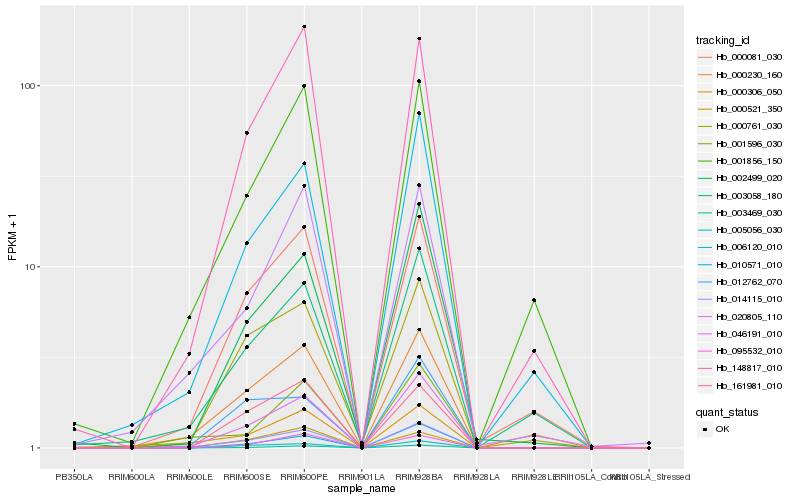
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_014115_010 |
0.0 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 2 |
Hb_046191_010 |
0.0554467084 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 3 |
Hb_000306_050 |
0.087328897 |
transcription factor |
TF Family: PLATZ |
protein with unknown function [Ricinus communis] |
| 4 |
Hb_005056_030 |
0.1034296327 |
- |
- |
DUF26 domain-containing protein 2 precursor, putative [Ricinus communis] |
| 5 |
Hb_000081_030 |
0.1106673412 |
- |
- |
BnaC09g05810D [Brassica napus] |
| 6 |
Hb_000761_030 |
0.1142915124 |
transcription factor |
TF Family: ERF |
Transcriptional factor TINY, putative [Ricinus communis] |
| 7 |
Hb_006120_010 |
0.1149490252 |
- |
- |
invertase inhibitor [Manihot esculenta] |
| 8 |
Hb_002499_020 |
0.1154268534 |
- |
- |
hevamine-A precursor, putative [Ricinus communis] |
| 9 |
Hb_003469_030 |
0.11743343 |
- |
- |
hypothetical protein JCGZ_01307 [Jatropha curcas] |
| 10 |
Hb_000230_160 |
0.1225790777 |
- |
- |
PREDICTED: uncharacterized protein LOC105631858 isoform X1 [Jatropha curcas] |
| 11 |
Hb_148817_010 |
0.124191638 |
- |
- |
PREDICTED: CASP-like protein 1D1 [Jatropha curcas] |
| 12 |
Hb_020805_110 |
0.1262533347 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 12 [Jatropha curcas] |
| 13 |
Hb_003058_180 |
0.1309373579 |
transcription factor |
TF Family: HSF |
hypothetical protein POPTR_0001s28040g [Populus trichocarpa] |
| 14 |
Hb_012762_070 |
0.133230538 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HOX3 [Jatropha curcas] |
| 15 |
Hb_010571_010 |
0.1332453215 |
- |
- |
PREDICTED: non-symbiotic hemoglobin 2 [Jatropha curcas] |
| 16 |
Hb_001856_150 |
0.1341560427 |
- |
- |
- |
| 17 |
Hb_001596_030 |
0.1377655266 |
- |
- |
PREDICTED: uncharacterized protein LOC105141672 [Populus euphratica] |
| 18 |
Hb_161981_010 |
0.1388201382 |
- |
- |
brassinosteroid insensitive 1 -like protein [Gossypium arboreum] |
| 19 |
Hb_000521_350 |
0.1394098664 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 20 |
Hb_095532_010 |
0.1409441799 |
- |
- |
BnaC05g27570D [Brassica napus] |