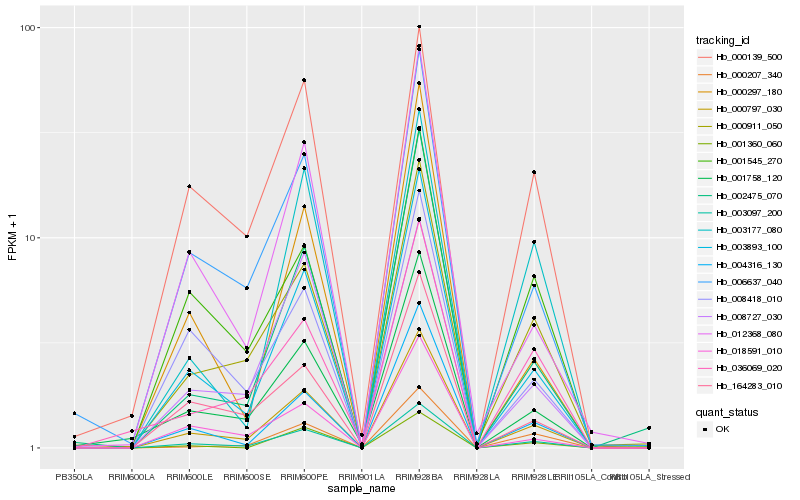
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003097_200 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000797_030 |
0.0211851062 |
- |
- |
PREDICTED: leucoanthocyanidin dioxygenase [Jatropha curcas] |
| 3 |
Hb_003893_100 |
0.060972693 |
- |
- |
PREDICTED: uncharacterized protein LOC105648774 [Jatropha curcas] |
| 4 |
Hb_000911_050 |
0.0938761097 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 21 [Jatropha curcas] |
| 5 |
Hb_006637_040 |
0.0997396271 |
- |
- |
phenylalanine ammonia-lyase 2 [Manihot esculenta] |
| 6 |
Hb_004316_130 |
0.1003769729 |
- |
- |
PREDICTED: glutathione transferase GST 23-like, partial [Eucalyptus grandis] |
| 7 |
Hb_001758_120 |
0.1116234527 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 8 |
Hb_012368_080 |
0.1142579228 |
- |
- |
PREDICTED: uncharacterized protein LOC105126277 [Populus euphratica] |
| 9 |
Hb_001545_270 |
0.1147709597 |
- |
- |
laccase, putative [Ricinus communis] |
| 10 |
Hb_008418_010 |
0.116862079 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH93-like [Jatropha curcas] |
| 11 |
Hb_164283_010 |
0.1181450608 |
- |
- |
hypothetical protein RCOM_0725500 [Ricinus communis] |
| 12 |
Hb_000207_340 |
0.1224294593 |
transcription factor |
TF Family: GRAS |
hypothetical protein POPTR_0013s11930g [Populus trichocarpa] |
| 13 |
Hb_018591_010 |
0.1235730805 |
- |
- |
kinase, putative [Ricinus communis] |
| 14 |
Hb_003177_080 |
0.1245449684 |
- |
- |
hypothetical protein L484_016032 [Morus notabilis] |
| 15 |
Hb_001360_060 |
0.1291540998 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 32 [Jatropha curcas] |
| 16 |
Hb_008727_030 |
0.1297211995 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000139_500 |
0.1321581765 |
- |
- |
PREDICTED: putative uncharacterized protein DDB_G0286901 [Prunus mume] |
| 18 |
Hb_036069_020 |
0.1322285538 |
- |
- |
PREDICTED: receptor protein kinase-like protein At4g34220 [Jatropha curcas] |
| 19 |
Hb_002475_070 |
0.1330799528 |
- |
- |
hypothetical protein POPTR_0017s02440g, partial [Populus trichocarpa] |
| 20 |
Hb_000297_180 |
0.1376921661 |
- |
- |
Desacetoxyvindoline 4-hydroxylase, putative [Ricinus communis] |