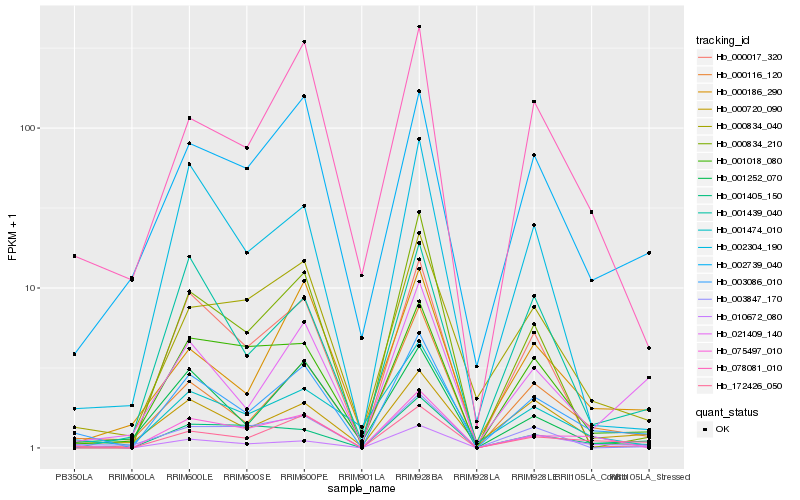
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003086_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_06093 [Jatropha curcas] |
| 2 |
Hb_172426_050 |
0.1145586628 |
- |
- |
PREDICTED: uncharacterized protein LOC105634266 [Jatropha curcas] |
| 3 |
Hb_000116_120 |
0.1479324701 |
- |
- |
PREDICTED: disease resistance response protein 206-like [Jatropha curcas] |
| 4 |
Hb_000720_090 |
0.1666756584 |
transcription factor |
TF Family: MYB |
myb, putative [Ricinus communis] |
| 5 |
Hb_021409_140 |
0.1703320781 |
- |
- |
PREDICTED: trehalase isoform X1 [Jatropha curcas] |
| 6 |
Hb_078081_010 |
0.1716360566 |
- |
- |
hypothetical protein PRUPE_ppa015726mg [Prunus persica] |
| 7 |
Hb_001405_150 |
0.1727488231 |
- |
- |
PREDICTED: protein At-4/1 [Jatropha curcas] |
| 8 |
Hb_001252_070 |
0.1771243525 |
- |
- |
hypothetical protein JCGZ_01385 [Jatropha curcas] |
| 9 |
Hb_002739_040 |
0.1801658543 |
- |
- |
PREDICTED: isocitrate dehydrogenase [NADP] [Jatropha curcas] |
| 10 |
Hb_075497_010 |
0.1828619491 |
- |
- |
hypothetical protein JCGZ_18511 [Jatropha curcas] |
| 11 |
Hb_010672_080 |
0.1834595127 |
transcription factor |
TF Family: MYB |
hypothetical protein JCGZ_00902 [Jatropha curcas] |
| 12 |
Hb_000834_040 |
0.1836706807 |
- |
- |
PREDICTED: ras-related protein Rab7 [Cucumis melo] |
| 13 |
Hb_001018_080 |
0.1844083027 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000186_290 |
0.1865910694 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 8 isoform X1 [Jatropha curcas] |
| 15 |
Hb_003847_170 |
0.1870826761 |
- |
- |
PREDICTED: trehalase isoform X1 [Jatropha curcas] |
| 16 |
Hb_000017_320 |
0.1876589447 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor GLABRA 3-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_000834_210 |
0.1885632255 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 18 |
Hb_001474_010 |
0.1907633322 |
- |
- |
PREDICTED: synaptotagmin-3 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001439_040 |
0.1909535504 |
- |
- |
PREDICTED: chorismate mutase 1, chloroplastic [Jatropha curcas] |
| 20 |
Hb_002304_190 |
0.1934201093 |
- |
- |
PREDICTED: 3'-N-debenzoyl-2'-deoxytaxol N-benzoyltransferase [Jatropha curcas] |