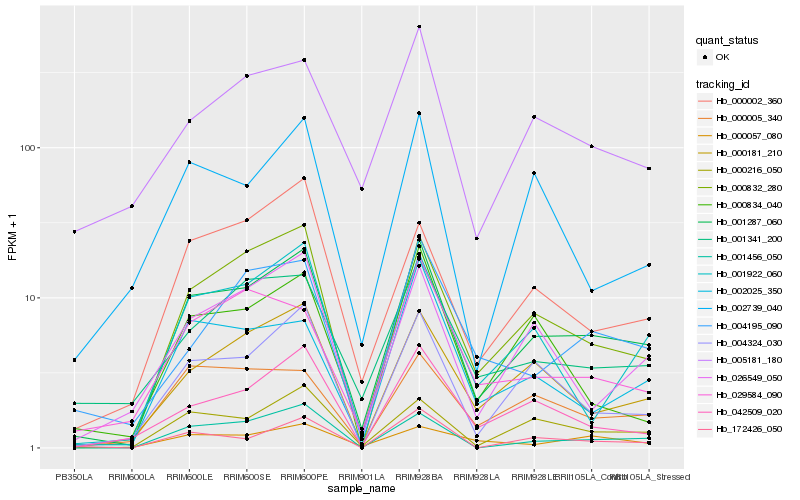
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001287_060 |
0.0 |
transcription factor |
TF Family: Orphans |
Salt-tolerance protein, putative [Ricinus communis] |
| 2 |
Hb_000832_280 |
0.1158535138 |
- |
- |
PREDICTED: aquaporin TIP1-3 [Jatropha curcas] |
| 3 |
Hb_000216_050 |
0.1265475582 |
- |
- |
PREDICTED: indole-3-acetaldehyde oxidase-like [Jatropha curcas] |
| 4 |
Hb_004324_030 |
0.1388145908 |
- |
- |
PREDICTED: probable protein phosphatase 2C 8 [Jatropha curcas] |
| 5 |
Hb_000181_210 |
0.1403413364 |
- |
- |
Phytosulfokine receptor precursor, putative [Ricinus communis] |
| 6 |
Hb_001456_050 |
0.1406804467 |
transcription factor |
TF Family: G2-like |
transcription factor, putative [Ricinus communis] |
| 7 |
Hb_001922_060 |
0.1516648685 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 8 |
Hb_042509_020 |
0.1571056532 |
- |
- |
hypothetical protein JCGZ_19555 [Jatropha curcas] |
| 9 |
Hb_000005_340 |
0.1582905134 |
- |
- |
PREDICTED: uncharacterized protein LOC105634028 isoform X1 [Jatropha curcas] |
| 10 |
Hb_172426_050 |
0.1583622582 |
- |
- |
PREDICTED: uncharacterized protein LOC105634266 [Jatropha curcas] |
| 11 |
Hb_026549_050 |
0.1585128489 |
- |
- |
PREDICTED: uncharacterized protein LOC105636246 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000002_360 |
0.1636509606 |
- |
- |
hypothetical protein POPTR_0008s16470g [Populus trichocarpa] |
| 13 |
Hb_001341_200 |
0.1647295009 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 4 [Jatropha curcas] |
| 14 |
Hb_005181_180 |
0.1671593541 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_004195_090 |
0.1717389881 |
- |
- |
PREDICTED: probable mitochondrial chaperone bcs1 [Jatropha curcas] |
| 16 |
Hb_002025_350 |
0.1753087768 |
- |
- |
PREDICTED: glutamate decarboxylase 4 [Jatropha curcas] |
| 17 |
Hb_000057_080 |
0.1759893205 |
- |
- |
PREDICTED: probable ATP-dependent RNA helicase DDX11 [Jatropha curcas] |
| 18 |
Hb_029584_090 |
0.1768452678 |
- |
- |
PREDICTED: inactive protein kinase SELMODRAFT_444075 [Jatropha curcas] |
| 19 |
Hb_000834_040 |
0.177588012 |
- |
- |
PREDICTED: ras-related protein Rab7 [Cucumis melo] |
| 20 |
Hb_002739_040 |
0.1783600306 |
- |
- |
PREDICTED: isocitrate dehydrogenase [NADP] [Jatropha curcas] |