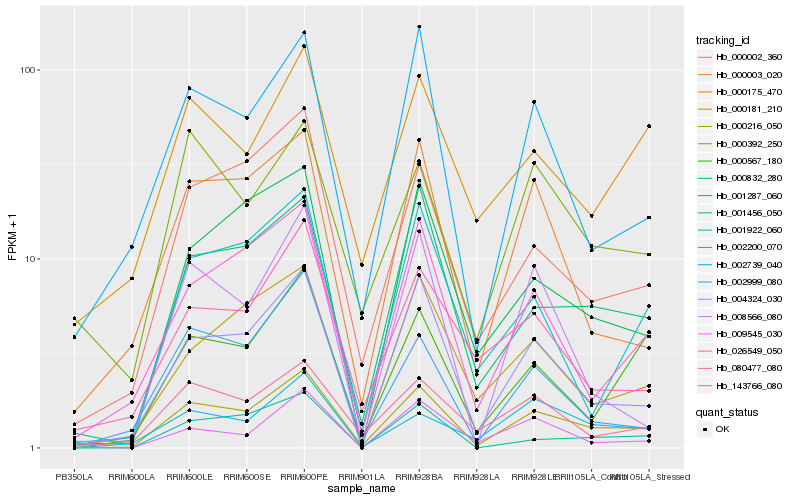
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000216_050 |
0.0 |
- |
- |
PREDICTED: indole-3-acetaldehyde oxidase-like [Jatropha curcas] |
| 2 |
Hb_004324_030 |
0.1162072223 |
- |
- |
PREDICTED: probable protein phosphatase 2C 8 [Jatropha curcas] |
| 3 |
Hb_001287_060 |
0.1265475582 |
transcription factor |
TF Family: Orphans |
Salt-tolerance protein, putative [Ricinus communis] |
| 4 |
Hb_000002_360 |
0.1466846414 |
- |
- |
hypothetical protein POPTR_0008s16470g [Populus trichocarpa] |
| 5 |
Hb_026549_050 |
0.1491326728 |
- |
- |
PREDICTED: uncharacterized protein LOC105636246 isoform X2 [Jatropha curcas] |
| 6 |
Hb_002739_040 |
0.1501920086 |
- |
- |
PREDICTED: isocitrate dehydrogenase [NADP] [Jatropha curcas] |
| 7 |
Hb_143766_080 |
0.1547236858 |
- |
- |
PREDICTED: uncharacterized protein LOC105637797 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000832_280 |
0.15730201 |
- |
- |
PREDICTED: aquaporin TIP1-3 [Jatropha curcas] |
| 9 |
Hb_000181_210 |
0.1582584127 |
- |
- |
Phytosulfokine receptor precursor, putative [Ricinus communis] |
| 10 |
Hb_000567_180 |
0.1587239176 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF1.5 [Jatropha curcas] |
| 11 |
Hb_000003_020 |
0.160186283 |
- |
- |
PREDICTED: uncharacterized protein LOC105631161 [Jatropha curcas] |
| 12 |
Hb_001456_050 |
0.1619243936 |
transcription factor |
TF Family: G2-like |
transcription factor, putative [Ricinus communis] |
| 13 |
Hb_001922_060 |
0.1622802167 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 14 |
Hb_000392_250 |
0.1689002381 |
- |
- |
PREDICTED: HVA22-like protein a [Jatropha curcas] |
| 15 |
Hb_002200_070 |
0.1689966962 |
- |
- |
PREDICTED: endoglucanase 9-like [Populus euphratica] |
| 16 |
Hb_002999_080 |
0.1696023086 |
transcription factor |
TF Family: G2-like |
PREDICTED: accessory gland protein Acp36DE [Jatropha curcas] |
| 17 |
Hb_000175_470 |
0.1729943826 |
- |
- |
lactoylglutathione lyase, putative [Ricinus communis] |
| 18 |
Hb_009545_030 |
0.1737470806 |
- |
- |
PREDICTED: folate-biopterin transporter 1, chloroplastic [Jatropha curcas] |
| 19 |
Hb_080477_080 |
0.1741854852 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4 isoform X1 [Gossypium raimondii] |
| 20 |
Hb_008566_080 |
0.1753237127 |
- |
- |
PREDICTED: uncharacterized protein LOC105647834 [Jatropha curcas] |