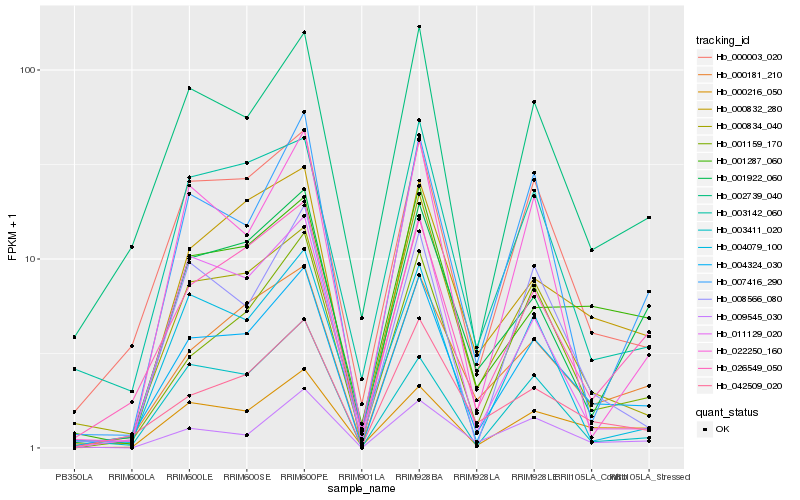
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004324_030 |
0.0 |
- |
- |
PREDICTED: probable protein phosphatase 2C 8 [Jatropha curcas] |
| 2 |
Hb_002739_040 |
0.1008697129 |
- |
- |
PREDICTED: isocitrate dehydrogenase [NADP] [Jatropha curcas] |
| 3 |
Hb_042509_020 |
0.1028647763 |
- |
- |
hypothetical protein JCGZ_19555 [Jatropha curcas] |
| 4 |
Hb_026549_050 |
0.1055529885 |
- |
- |
PREDICTED: uncharacterized protein LOC105636246 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000003_020 |
0.1088578639 |
- |
- |
PREDICTED: uncharacterized protein LOC105631161 [Jatropha curcas] |
| 6 |
Hb_000181_210 |
0.1092843001 |
- |
- |
Phytosulfokine receptor precursor, putative [Ricinus communis] |
| 7 |
Hb_000216_050 |
0.1162072223 |
- |
- |
PREDICTED: indole-3-acetaldehyde oxidase-like [Jatropha curcas] |
| 8 |
Hb_001159_170 |
0.1228391663 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 1 [Jatropha curcas] |
| 9 |
Hb_000832_280 |
0.126784184 |
- |
- |
PREDICTED: aquaporin TIP1-3 [Jatropha curcas] |
| 10 |
Hb_000834_040 |
0.1297257969 |
- |
- |
PREDICTED: ras-related protein Rab7 [Cucumis melo] |
| 11 |
Hb_007416_290 |
0.1359656035 |
- |
- |
PREDICTED: plasma membrane ATPase 4-like [Pyrus x bretschneideri] |
| 12 |
Hb_009545_030 |
0.1365779702 |
- |
- |
PREDICTED: folate-biopterin transporter 1, chloroplastic [Jatropha curcas] |
| 13 |
Hb_001287_060 |
0.1388145908 |
transcription factor |
TF Family: Orphans |
Salt-tolerance protein, putative [Ricinus communis] |
| 14 |
Hb_003142_060 |
0.1390738633 |
- |
- |
PREDICTED: glutamate synthase 1 [NADH], chloroplastic isoform X1 [Jatropha curcas] |
| 15 |
Hb_008566_080 |
0.1401368125 |
- |
- |
PREDICTED: uncharacterized protein LOC105647834 [Jatropha curcas] |
| 16 |
Hb_001922_060 |
0.1415188191 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 17 |
Hb_022250_160 |
0.1485324377 |
- |
- |
respiratory burst oxidase A [Manihot esculenta] |
| 18 |
Hb_004079_100 |
0.1495416564 |
- |
- |
PREDICTED: uncharacterized protein LOC105637705 [Jatropha curcas] |
| 19 |
Hb_011129_020 |
0.1504249649 |
- |
- |
Beta-1,3-galactosyltransferase sqv-2, putative [Ricinus communis] |
| 20 |
Hb_003411_020 |
0.1505066241 |
- |
- |
alpha-glucosidase, putative [Ricinus communis] |