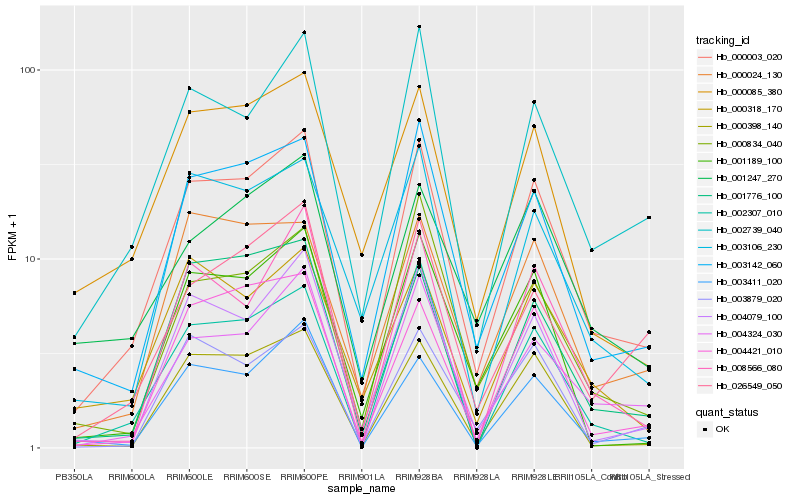
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000003_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105631161 [Jatropha curcas] |
| 2 |
Hb_003142_060 |
0.0895589904 |
- |
- |
PREDICTED: glutamate synthase 1 [NADH], chloroplastic isoform X1 [Jatropha curcas] |
| 3 |
Hb_002739_040 |
0.1000763298 |
- |
- |
PREDICTED: isocitrate dehydrogenase [NADP] [Jatropha curcas] |
| 4 |
Hb_004324_030 |
0.1088578639 |
- |
- |
PREDICTED: probable protein phosphatase 2C 8 [Jatropha curcas] |
| 5 |
Hb_004079_100 |
0.1202339709 |
- |
- |
PREDICTED: uncharacterized protein LOC105637705 [Jatropha curcas] |
| 6 |
Hb_000085_380 |
0.1213845875 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_003106_230 |
0.1217103407 |
transcription factor |
TF Family: LIM |
PREDICTED: LIM domain-containing protein WLIM1-like [Jatropha curcas] |
| 8 |
Hb_003411_020 |
0.1217817509 |
- |
- |
alpha-glucosidase, putative [Ricinus communis] |
| 9 |
Hb_000398_140 |
0.122462155 |
- |
- |
PREDICTED: methionine S-methyltransferase [Jatropha curcas] |
| 10 |
Hb_004421_010 |
0.1250394886 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 11 |
Hb_000024_130 |
0.126629478 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000318_170 |
0.1275384553 |
- |
- |
N-acetyl-gamma-glutamyl-phosphate reductase, putative [Ricinus communis] |
| 13 |
Hb_002307_010 |
0.1276351567 |
- |
- |
PREDICTED: uncharacterized protein LOC105632092 [Jatropha curcas] |
| 14 |
Hb_026549_050 |
0.1281492604 |
- |
- |
PREDICTED: uncharacterized protein LOC105636246 isoform X2 [Jatropha curcas] |
| 15 |
Hb_001189_100 |
0.1310265152 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 2-like [Jatropha curcas] |
| 16 |
Hb_003879_020 |
0.1310786591 |
- |
- |
PREDICTED: MATE efflux family protein 1-like [Jatropha curcas] |
| 17 |
Hb_001776_100 |
0.1333761233 |
- |
- |
PREDICTED: outer envelope pore protein 16, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000834_040 |
0.1335926797 |
- |
- |
PREDICTED: ras-related protein Rab7 [Cucumis melo] |
| 19 |
Hb_001247_270 |
0.1361339644 |
- |
- |
PREDICTED: probable L-cysteine desulfhydrase, chloroplastic [Jatropha curcas] |
| 20 |
Hb_008566_080 |
0.1365081036 |
- |
- |
PREDICTED: uncharacterized protein LOC105647834 [Jatropha curcas] |