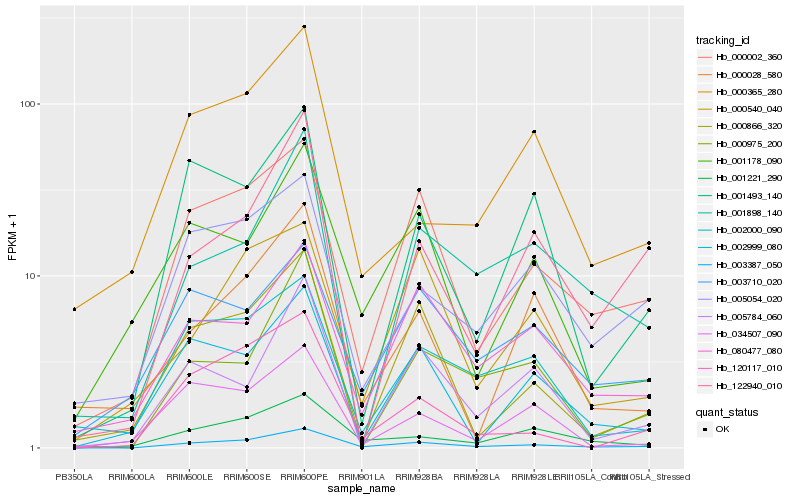
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003387_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105639324 [Jatropha curcas] |
| 2 |
Hb_000002_360 |
0.1318412914 |
- |
- |
hypothetical protein POPTR_0008s16470g [Populus trichocarpa] |
| 3 |
Hb_001221_290 |
0.1543722635 |
- |
- |
hypothetical protein RCOM_1506010 [Ricinus communis] |
| 4 |
Hb_080477_080 |
0.1723215449 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4 isoform X1 [Gossypium raimondii] |
| 5 |
Hb_005054_020 |
0.1740316044 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g11320 [Jatropha curcas] |
| 6 |
Hb_000866_320 |
0.1824180519 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 7 |
Hb_120117_010 |
0.1825943961 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 8 |
Hb_122940_010 |
0.1840899788 |
- |
- |
PREDICTED: 2-isopropylmalate synthase 2, chloroplastic-like [Populus euphratica] |
| 9 |
Hb_000975_200 |
0.1865885389 |
- |
- |
PREDICTED: uncharacterized protein LOC105628231 isoform X2 [Jatropha curcas] |
| 10 |
Hb_002999_080 |
0.1884820877 |
transcription factor |
TF Family: G2-like |
PREDICTED: accessory gland protein Acp36DE [Jatropha curcas] |
| 11 |
Hb_001898_140 |
0.1885345508 |
- |
- |
hypothetical protein AMTR_s00071p00184460 [Amborella trichopoda] |
| 12 |
Hb_002000_090 |
0.1896260208 |
- |
- |
60S ribosomal protein L3B [Hevea brasiliensis] |
| 13 |
Hb_001493_140 |
0.1900588991 |
- |
- |
hypothetical protein PHAVU_010G123100g [Phaseolus vulgaris] |
| 14 |
Hb_003710_020 |
0.1903919513 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g53440 isoform X1 [Jatropha curcas] |
| 15 |
Hb_005784_060 |
0.1958833382 |
- |
- |
PREDICTED: U-box domain-containing protein 28 [Jatropha curcas] |
| 16 |
Hb_000028_580 |
0.1962875728 |
- |
- |
PREDICTED: glutamate decarboxylase 5-like [Jatropha curcas] |
| 17 |
Hb_000365_280 |
0.1971753874 |
- |
- |
calcium binding protein/cast, putative [Ricinus communis] |
| 18 |
Hb_001178_090 |
0.1977010589 |
- |
- |
Monodehydroascorbate reductase family protein [Populus trichocarpa] |
| 19 |
Hb_000540_040 |
0.1994283962 |
- |
- |
PREDICTED: chitinase 2 [Amborella trichopoda] |
| 20 |
Hb_034507_090 |
0.2004356335 |
- |
- |
PREDICTED: UPF0503 protein At3g09070, chloroplastic [Jatropha curcas] |