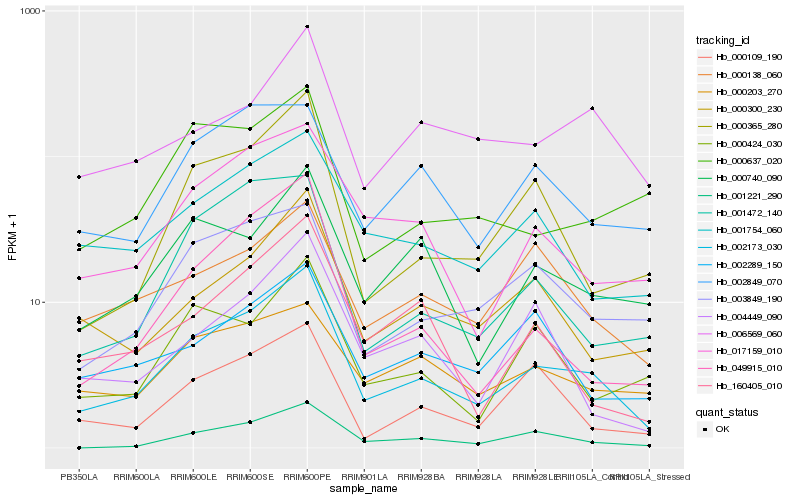
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002173_030 |
0.0 |
- |
- |
hypothetical protein JCGZ_12466 [Jatropha curcas] |
| 2 |
Hb_000365_280 |
0.1485490355 |
- |
- |
calcium binding protein/cast, putative [Ricinus communis] |
| 3 |
Hb_001221_290 |
0.1517539546 |
- |
- |
hypothetical protein RCOM_1506010 [Ricinus communis] |
| 4 |
Hb_017159_010 |
0.1560665598 |
- |
- |
- |
| 5 |
Hb_049915_010 |
0.1576448116 |
- |
- |
PREDICTED: uncharacterized serine-rich protein C215.13 [Jatropha curcas] |
| 6 |
Hb_000300_230 |
0.1622732908 |
- |
- |
PREDICTED: cation/calcium exchanger 2 [Jatropha curcas] |
| 7 |
Hb_001754_060 |
0.1625191283 |
- |
- |
Xyloglucan 6-xylosyltransferase, putative [Ricinus communis] |
| 8 |
Hb_003849_190 |
0.1639817815 |
- |
- |
PREDICTED: uncharacterized protein LOC105644700 [Jatropha curcas] |
| 9 |
Hb_001472_140 |
0.1646370059 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g41330 [Jatropha curcas] |
| 10 |
Hb_000109_190 |
0.1701150394 |
- |
- |
PREDICTED: RING-H2 finger protein ATL47 [Jatropha curcas] |
| 11 |
Hb_000740_090 |
0.1724444653 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_160405_010 |
0.1727532306 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 13 |
Hb_000424_030 |
0.1785377945 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000138_060 |
0.1812603768 |
- |
- |
PREDICTED: protein trichome birefringence [Jatropha curcas] |
| 15 |
Hb_000637_020 |
0.1820672306 |
- |
- |
PREDICTED: uncharacterized protein LOC105650669 [Jatropha curcas] |
| 16 |
Hb_004449_090 |
0.1821718568 |
- |
- |
PREDICTED: CO(2)-response secreted protease-like [Jatropha curcas] |
| 17 |
Hb_002289_150 |
0.1828121801 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 3 [UDP-forming]-like [Jatropha curcas] |
| 18 |
Hb_002849_070 |
0.1840488666 |
- |
- |
PREDICTED: protein BPS1, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000203_270 |
0.1858228223 |
- |
- |
alpha-xylosidase, putative [Ricinus communis] |
| 20 |
Hb_006569_060 |
0.1865049579 |
- |
- |
plasma membrane intrinsic protein PIP2;3 [Hevea brasiliensis] |