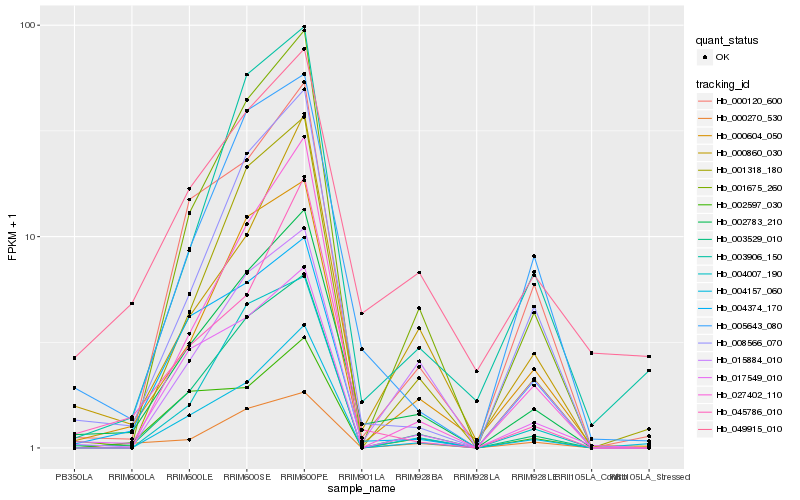
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002783_210 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105635341 [Jatropha curcas] |
| 2 |
Hb_003529_010 |
0.1063871114 |
- |
- |
PREDICTED: disease resistance protein RPS6-like [Jatropha curcas] |
| 3 |
Hb_001675_260 |
0.1169842492 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 4 |
Hb_000604_050 |
0.1210489833 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 8-like [Jatropha curcas] |
| 5 |
Hb_008566_070 |
0.1238657012 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_017549_010 |
0.1281052423 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_00222 [Jatropha curcas] |
| 7 |
Hb_005643_080 |
0.1339013574 |
- |
- |
hypothetical protein POPTR_0001s12740g [Populus trichocarpa] |
| 8 |
Hb_000120_600 |
0.1353576702 |
- |
- |
PREDICTED: uncharacterized protein LOC105628582 [Jatropha curcas] |
| 9 |
Hb_004157_060 |
0.1491784047 |
- |
- |
PREDICTED: xyloglucan endotransglucosylase/hydrolase protein 22-like [Jatropha curcas] |
| 10 |
Hb_004374_170 |
0.150207802 |
- |
- |
PREDICTED: gibberellin 20 oxidase 1-B [Jatropha curcas] |
| 11 |
Hb_001318_180 |
0.1505932469 |
- |
- |
PREDICTED: protein NIM1-INTERACTING 1 [Jatropha curcas] |
| 12 |
Hb_000860_030 |
0.1508505765 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_003906_150 |
0.151465778 |
- |
- |
peptidoglycan-binding LysM domain-containing family protein, partial [Populus trichocarpa] |
| 14 |
Hb_027402_110 |
0.1514712123 |
- |
- |
PREDICTED: uncharacterized protein LOC105640545 [Jatropha curcas] |
| 15 |
Hb_045786_010 |
0.1520686803 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 16 |
Hb_000270_530 |
0.1549836438 |
- |
- |
hypothetical protein POPTR_0015s08220g [Populus trichocarpa] |
| 17 |
Hb_049915_010 |
0.1564428755 |
- |
- |
PREDICTED: uncharacterized serine-rich protein C215.13 [Jatropha curcas] |
| 18 |
Hb_004007_190 |
0.1575675807 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein AZF2-like [Jatropha curcas] |
| 19 |
Hb_002597_030 |
0.1623247805 |
- |
- |
- |
| 20 |
Hb_015884_010 |
0.163595726 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g63930 isoform X1 [Jatropha curcas] |