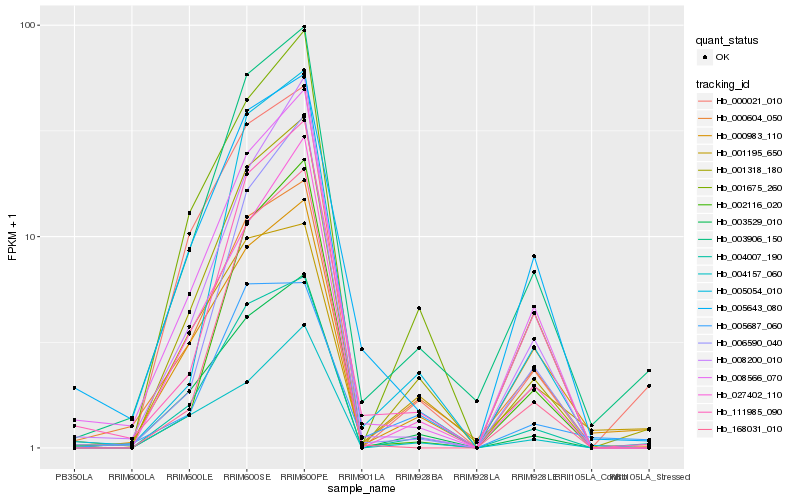
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003906_150 |
0.0 |
- |
- |
peptidoglycan-binding LysM domain-containing family protein, partial [Populus trichocarpa] |
| 2 |
Hb_000604_050 |
0.0898171352 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 8-like [Jatropha curcas] |
| 3 |
Hb_001318_180 |
0.0965173604 |
- |
- |
PREDICTED: protein NIM1-INTERACTING 1 [Jatropha curcas] |
| 4 |
Hb_008566_070 |
0.1087756714 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_111985_090 |
0.111151905 |
- |
- |
hypothetical protein RCOM_1595950 [Ricinus communis] |
| 6 |
Hb_004007_190 |
0.1113958681 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein AZF2-like [Jatropha curcas] |
| 7 |
Hb_000983_110 |
0.120592867 |
- |
- |
PREDICTED: omega-hydroxypalmitate O-feruloyl transferase [Jatropha curcas] |
| 8 |
Hb_001675_260 |
0.1289131351 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 9 |
Hb_003529_010 |
0.1306250367 |
- |
- |
PREDICTED: disease resistance protein RPS6-like [Jatropha curcas] |
| 10 |
Hb_005054_010 |
0.1315711833 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 11 |
Hb_000021_010 |
0.1322110528 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_005643_080 |
0.133131651 |
- |
- |
hypothetical protein POPTR_0001s12740g [Populus trichocarpa] |
| 13 |
Hb_027402_110 |
0.1400975452 |
- |
- |
PREDICTED: uncharacterized protein LOC105640545 [Jatropha curcas] |
| 14 |
Hb_006590_040 |
0.1428124346 |
- |
- |
amino acid permease, putative [Ricinus communis] |
| 15 |
Hb_002116_020 |
0.1433103139 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 35 [Jatropha curcas] |
| 16 |
Hb_008200_010 |
0.1436712983 |
- |
- |
PREDICTED: ABC transporter G family member 14-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_168031_010 |
0.1460514866 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.6 [Jatropha curcas] |
| 18 |
Hb_001195_650 |
0.1470666061 |
- |
- |
PREDICTED: calcium uniporter protein 4, mitochondrial [Jatropha curcas] |
| 19 |
Hb_004157_060 |
0.1472900849 |
- |
- |
PREDICTED: xyloglucan endotransglucosylase/hydrolase protein 22-like [Jatropha curcas] |
| 20 |
Hb_005687_060 |
0.1476652163 |
- |
- |
PREDICTED: cinnamoyl-CoA reductase 1-like isoform X1 [Jatropha curcas] |