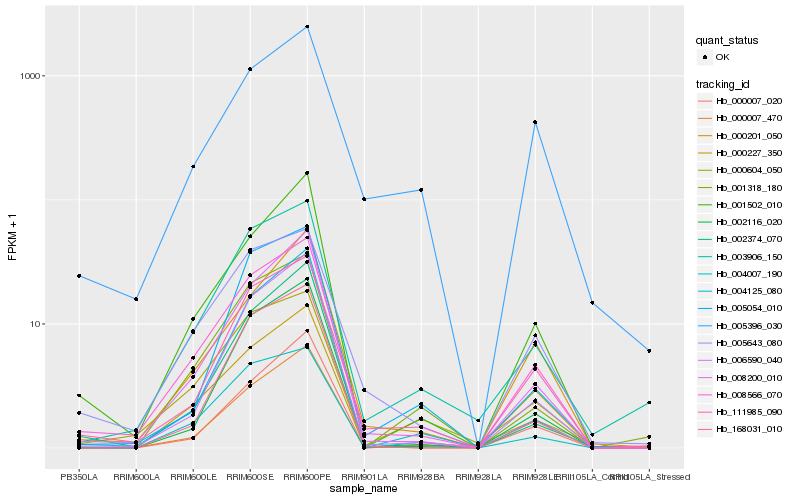
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_111985_090 |
0.0 |
- |
- |
hypothetical protein RCOM_1595950 [Ricinus communis] |
| 2 |
Hb_008566_070 |
0.0891412004 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_003906_150 |
0.111151905 |
- |
- |
peptidoglycan-binding LysM domain-containing family protein, partial [Populus trichocarpa] |
| 4 |
Hb_006590_040 |
0.1118095311 |
- |
- |
amino acid permease, putative [Ricinus communis] |
| 5 |
Hb_005054_010 |
0.112507686 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 6 |
Hb_002116_020 |
0.113144952 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 35 [Jatropha curcas] |
| 7 |
Hb_005643_080 |
0.1201066503 |
- |
- |
hypothetical protein POPTR_0001s12740g [Populus trichocarpa] |
| 8 |
Hb_008200_010 |
0.1252535152 |
- |
- |
PREDICTED: ABC transporter G family member 14-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_168031_010 |
0.1300394261 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.6 [Jatropha curcas] |
| 10 |
Hb_005396_030 |
0.1311703921 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000604_050 |
0.1335871151 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 8-like [Jatropha curcas] |
| 12 |
Hb_000201_050 |
0.1352207745 |
- |
- |
- |
| 13 |
Hb_000007_020 |
0.1379626448 |
transcription factor |
TF Family: bZIP |
Ocs element-binding factor, putative [Ricinus communis] |
| 14 |
Hb_000227_350 |
0.1400909436 |
- |
- |
hypothetical protein CICLE_v10006537mg [Citrus clementina] |
| 15 |
Hb_000007_470 |
0.1407231736 |
- |
- |
PREDICTED: phosphate transporter PHO1 homolog 1 [Jatropha curcas] |
| 16 |
Hb_001502_010 |
0.1426036323 |
- |
- |
hypothetical protein JCGZ_04288 [Jatropha curcas] |
| 17 |
Hb_004125_080 |
0.1449668818 |
- |
- |
PREDICTED: dCTP pyrophosphatase 1-like [Populus euphratica] |
| 18 |
Hb_004007_190 |
0.1473377314 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein AZF2-like [Jatropha curcas] |
| 19 |
Hb_001318_180 |
0.1492983568 |
- |
- |
PREDICTED: protein NIM1-INTERACTING 1 [Jatropha curcas] |
| 20 |
Hb_002374_070 |
0.1502388535 |
- |
- |
amino acid transporter, putative [Ricinus communis] |