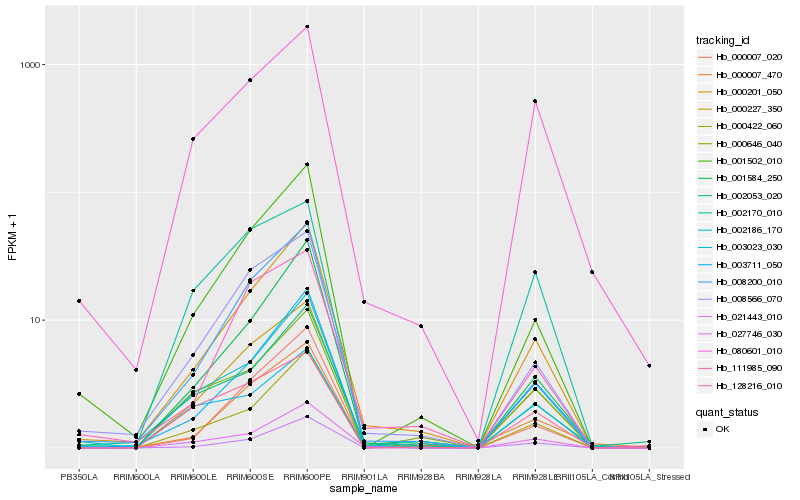
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000227_350 |
0.0 |
- |
- |
hypothetical protein CICLE_v10006537mg [Citrus clementina] |
| 2 |
Hb_000201_050 |
0.104212512 |
- |
- |
- |
| 3 |
Hb_008566_070 |
0.1048181551 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002170_010 |
0.1136373273 |
- |
- |
hypothetical protein JCGZ_25046 [Jatropha curcas] |
| 5 |
Hb_002186_170 |
0.1152157457 |
- |
- |
hypothetical protein POPTR_0067s00200g [Populus trichocarpa] |
| 6 |
Hb_128216_010 |
0.1153519066 |
- |
- |
PREDICTED: UDP-glycosyltransferase 79B6-like [Jatropha curcas] |
| 7 |
Hb_080601_010 |
0.119320289 |
- |
- |
metallothionein 3-like protein [Hevea brasiliensis] |
| 8 |
Hb_027746_030 |
0.1193500589 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_003711_050 |
0.1217095226 |
- |
- |
Isoflavone reductase, putative [Ricinus communis] |
| 10 |
Hb_001502_010 |
0.1273533738 |
- |
- |
hypothetical protein JCGZ_04288 [Jatropha curcas] |
| 11 |
Hb_008200_010 |
0.1317632336 |
- |
- |
PREDICTED: ABC transporter G family member 14-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_000007_470 |
0.133736456 |
- |
- |
PREDICTED: phosphate transporter PHO1 homolog 1 [Jatropha curcas] |
| 13 |
Hb_000646_040 |
0.1339257072 |
- |
- |
PREDICTED: thiamine-repressible mitochondrial transport protein THI74-like [Jatropha curcas] |
| 14 |
Hb_021443_010 |
0.1341210005 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 1 [Jatropha curcas] |
| 15 |
Hb_000422_060 |
0.1390476299 |
transcription factor |
TF Family: OFP |
hypothetical protein RCOM_1282480 [Ricinus communis] |
| 16 |
Hb_111985_090 |
0.1400909436 |
- |
- |
hypothetical protein RCOM_1595950 [Ricinus communis] |
| 17 |
Hb_000007_020 |
0.1412111278 |
transcription factor |
TF Family: bZIP |
Ocs element-binding factor, putative [Ricinus communis] |
| 18 |
Hb_003023_030 |
0.141727925 |
- |
- |
PREDICTED: MATE efflux family protein 5-like isoform X1 [Citrus sinensis] |
| 19 |
Hb_002053_020 |
0.1427816919 |
- |
- |
PREDICTED: MATE efflux family protein DTX1 [Jatropha curcas] |
| 20 |
Hb_001584_250 |
0.143929051 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |