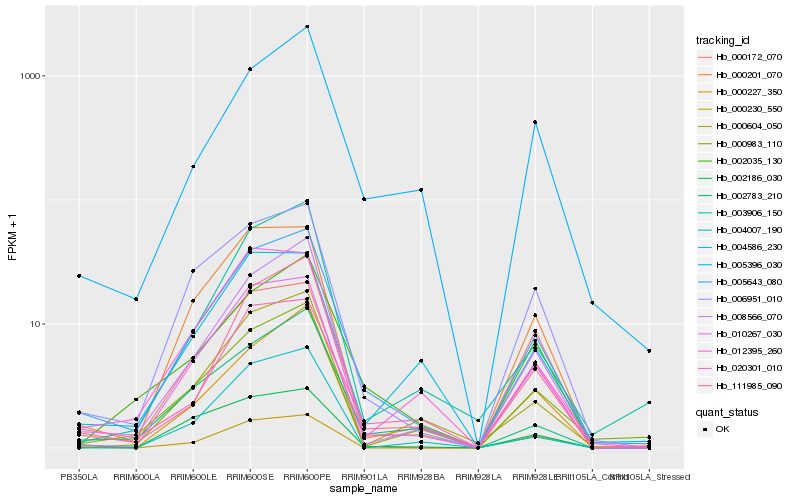
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005643_080 |
0.0 |
- |
- |
hypothetical protein POPTR_0001s12740g [Populus trichocarpa] |
| 2 |
Hb_008566_070 |
0.1006255552 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_006951_010 |
0.110814246 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 10-like [Populus euphratica] |
| 4 |
Hb_111985_090 |
0.1201066503 |
- |
- |
hypothetical protein RCOM_1595950 [Ricinus communis] |
| 5 |
Hb_000604_050 |
0.1231211512 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 8-like [Jatropha curcas] |
| 6 |
Hb_000201_070 |
0.1244276158 |
- |
- |
hypothetical protein POPTR_0067s00200g [Populus trichocarpa] |
| 7 |
Hb_010267_030 |
0.1244375544 |
- |
- |
DUF26 domain-containing protein 1 precursor, putative [Ricinus communis] |
| 8 |
Hb_020301_010 |
0.1281379416 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 9 |
Hb_002035_130 |
0.1318349893 |
- |
- |
plasma membrane H+-ATPase [Hevea brasiliensis] |
| 10 |
Hb_005396_030 |
0.1321694898 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_003906_150 |
0.133131651 |
- |
- |
peptidoglycan-binding LysM domain-containing family protein, partial [Populus trichocarpa] |
| 12 |
Hb_002783_210 |
0.1339013574 |
- |
- |
PREDICTED: uncharacterized protein LOC105635341 [Jatropha curcas] |
| 13 |
Hb_000983_110 |
0.136245593 |
- |
- |
PREDICTED: omega-hydroxypalmitate O-feruloyl transferase [Jatropha curcas] |
| 14 |
Hb_002186_030 |
0.1445684693 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g53430 isoform X1 [Populus euphratica] |
| 15 |
Hb_012395_260 |
0.1467212691 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA13 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000227_350 |
0.1511623219 |
- |
- |
hypothetical protein CICLE_v10006537mg [Citrus clementina] |
| 17 |
Hb_004586_230 |
0.1526450078 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000172_070 |
0.1533890898 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 19 |
Hb_000230_550 |
0.1548286945 |
- |
- |
PREDICTED: adenylate isopentenyltransferase 5, chloroplastic [Jatropha curcas] |
| 20 |
Hb_004007_190 |
0.1552579314 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein AZF2-like [Jatropha curcas] |