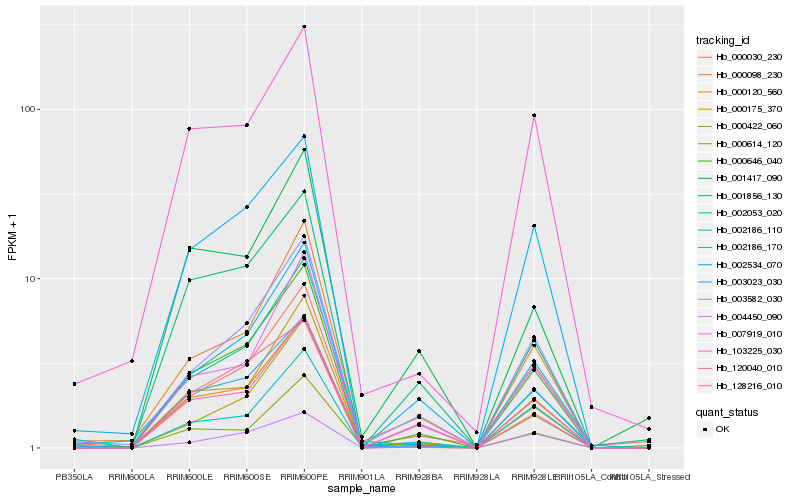
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000646_040 |
0.0 |
- |
- |
PREDICTED: thiamine-repressible mitochondrial transport protein THI74-like [Jatropha curcas] |
| 2 |
Hb_000120_560 |
0.0740263225 |
- |
- |
protein with unknown function [Ricinus communis] |
| 3 |
Hb_003582_030 |
0.0828736428 |
- |
- |
PREDICTED: serine/threonine-protein kinase BRI1-like 2 [Jatropha curcas] |
| 4 |
Hb_002534_070 |
0.0926737052 |
- |
- |
PREDICTED: uncharacterized protein LOC105640545 [Jatropha curcas] |
| 5 |
Hb_007919_010 |
0.0939867832 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002186_170 |
0.096308569 |
- |
- |
hypothetical protein POPTR_0067s00200g [Populus trichocarpa] |
| 7 |
Hb_000422_060 |
0.0968675503 |
transcription factor |
TF Family: OFP |
hypothetical protein RCOM_1282480 [Ricinus communis] |
| 8 |
Hb_000098_230 |
0.0972927259 |
- |
- |
Potassium channel AKT2/3, putative [Ricinus communis] |
| 9 |
Hb_003023_030 |
0.0996025619 |
- |
- |
PREDICTED: MATE efflux family protein 5-like isoform X1 [Citrus sinensis] |
| 10 |
Hb_000175_370 |
0.1075766262 |
- |
- |
hypothetical protein EUGRSUZ_F03862 [Eucalyptus grandis] |
| 11 |
Hb_000030_230 |
0.1094858124 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme [Jatropha curcas] |
| 12 |
Hb_120040_010 |
0.1169799235 |
- |
- |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 13 |
Hb_002186_110 |
0.1174618624 |
- |
- |
hypothetical protein CICLE_v10024045mg, partial [Citrus clementina] |
| 14 |
Hb_002053_020 |
0.1186591393 |
- |
- |
PREDICTED: MATE efflux family protein DTX1 [Jatropha curcas] |
| 15 |
Hb_128216_010 |
0.1198304026 |
- |
- |
PREDICTED: UDP-glycosyltransferase 79B6-like [Jatropha curcas] |
| 16 |
Hb_000614_120 |
0.1201265713 |
- |
- |
hypothetical protein JCGZ_17923 [Jatropha curcas] |
| 17 |
Hb_001856_130 |
0.1202326429 |
- |
- |
hypothetical protein CISIN_1g022795mg [Citrus sinensis] |
| 18 |
Hb_004450_090 |
0.1215185788 |
- |
- |
TRANSPARENT TESTA 12 protein, putative [Ricinus communis] |
| 19 |
Hb_103225_030 |
0.1227526176 |
transcription factor |
TF Family: Orphans |
DNA binding protein, putative [Ricinus communis] |
| 20 |
Hb_001417_090 |
0.122795298 |
- |
- |
PREDICTED: uncharacterized protein LOC105650625 [Jatropha curcas] |